- CSE254, Intrinsic Dimension 2019
- Geometric Computation group in the Computer Science Department of Stanford University.
- CSIC 5011: Topological and Geometric Data Reduction and Visualization Fall 2019
Principal component analysis or singular value decomposition can be applied to matrix approximation. The data collected in practice always save in the table form, which can considered as a matrix. Another techniques similar to PCA is eigenvalue-eigenvector decomposition. Dimension reduction is really the topic of data science as data preprocessing .
The basic idea of dimension reduction is that not all information is necessary for a specific task.
The motivation of dimension reduction is Curse of Dimensionality, limit of storage/computation and so on. The high dimensional space is not easy for us to visualize, imagine or understand. The intuition or insight to high dimensional space is weak for us, the people who live in the three dimensional space.
As a preprocessing data method, it helps us to select features and learn proper representation.
The dimension reduction is related with geometry of data set, which includes manifold learning and topological data analysis.
All manifold learning algorithms assume that data set lies on a smooth non-linear manifold of low dimension and a mapping
(where
It is a wonderful review of dimension reduction at TiCC TR 2009–005, Dimensionality Reduction: A Comparative Review, Laurens van der Maaten Eric Postma, Jaap van den Herik TiCC, Tilburg University.
A related top is data compression, a branch of information theory, a more useful and fundamental topic in computer science.
- https://lvdmaaten.github.io/software/
- https://jakevdp.github.io/PythonDataScienceHandbook/05.10-manifold-learning.html
- http://www.lcayton.com/resexam.pdf
- https://www.wikiwand.com/en/Data_compression
- https://brilliant.org/wiki/compression/
- https://www.wikiwand.com/en/Curse_of_dimensionality
- http://www.cnblogs.com/xbinworld/archive/2012/07/09/LLE.html
- https://www.ayasdi.com/blog/artificial-intelligence/prediction-needs-unsupervised-learning/
- Nonlinear Dimensionality Reduction
- COMPSCI 630: Randomized Algorithms
- Dimensionality reduction
- Surprises in high dimensions
- Linear dimension reduction via Johnson-Lindenstrauss
- manifold learning with applications to object recognition
- http://bactra.org/notebooks/manifold-learning.html
- http://www.cs.columbia.edu/~jebara/papers/BlakeShawFinalThesis.pdf
- https://www.zhihu.com/question/41106133
The data in table form can be regarded as matrix in mathematics. And we can apply singular value decomposition to low rank approximation or non-negative matrix factorization, which we will talk in PCA and SVD. It is classified as linear techniques. And it can extend to kernel PCA and generalized PCA.
Multi-Dimension Scaling is a distance-preserving manifold learning method. Distance preserving methods assume that a manifold can be defined by the pairwise distances of its points. In distance preserving methods, a low dimensional embedding is obtained from the higher dimension in such a way that pairwise distances between the points remain same. Some distance preserving methods preserve spatial distances (MDS) while some preserve graph distances.
MDS is not a single method but a family of methods. MDS takes a dissimilarity or distance matrix
It is a linear map $$ {X}\in\mathbb{R}^D\to {Z}\in\mathbb{R}^d\ Z = W^T X $$
Steps of a Classical MDS algorithm:
Classical MDS uses the fact that the coordinate matrix can be derived by eigenvalue decomposition from
- Set up the squared proximity matrix
${\textstyle D^{(2)}=[d_{ij}^{2}]}$ - Apply double centering:
$B=-{\frac{1}{2}J D^{(2)}J}$ using the centering matrix${\textstyle J=I-{\frac {1}{n}11'}}$ , where${\textstyle n}$ is the number of objects. - Determine the
${\textstyle m}$ largest eigenvalues$\lambda_{1},\lambda_{2},...,\lambda_{m}$ and corresponding eigenvectors${\textstyle e_{1},e_{2},...,e_{m}}$ of${\textstyle B}$ (where${\textstyle m}$ is the number of dimensions desired for the output). - Now,
${\textstyle Z=E_{m}\Lambda_{m}^{1/2}}$ , where${\textstyle E_{m}}$ is the matrix of${\textstyle m}$ eigenvectors and${\textstyle \Lambda_{m}}$ is the diagonal matrix of${\textstyle m}$ eigenvalues of${\textstyle B}$ .
Classical MDS assumes Euclidean distances. So this is not applicable for direct dissimilarity ratings.
- http://www.math.pku.edu.cn/teachers/yaoy/reference/book05.pdf
- http://www.statsoft.com/textbook/multidimensional-scaling
- https://www.springer.com/in/book/9780387251509
- https://www.stat.pitt.edu/sungkyu/course/2221Fall13/lec8_mds_combined.pdf
- https://www.stat.pitt.edu/sungkyu/course/2221Fall13/lec4_pca_slides.pdf
- https://www.ibm.com/support/knowledgecenter/en/SSLVMB_22.0.0/
- https://www.wikiwand.com/en/Multidimensional_scaling
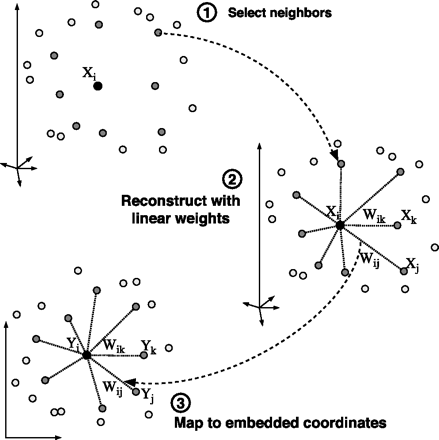
Locally Linear Embedding(LLE) is a topology preserving manifold learning method. Topology preservation means the neighborhood structure is intact. Methods like SOM(self-organizing map) are also topology preserving but they assume a predefined lattice for the lower manifold. LLE creates the lattice based on the information contained in the dataset.
- Compute the neighbors of each data point,
$\vec{X}_i$ . - Compute the weights
$W_{ij}$ that best reconstruct each data point$\vec{x_i}$ from its neighbors, minimizing the cost $\sum_{i}|\vec{X}i - \sum{j}W_{ij}\vec{X}_j|^2$ by constrained linear fits. - Compute the vectors $\vec{Y}i$ best reconstructed by the weights $W{ij}$, minimizing the quadratic form $\sum_{i}|\vec{Y}i - \sum{j}W_{ij}\vec{Y}_j|^2$ by its bottom nonzero eigenvectors.
- https://cs.nyu.edu/~roweis/lle/
- http://www.robots.ox.ac.uk/~az/lectures/ml/lle.pdf
- http://ai.stanford.edu/~schuon/learning/inclle.pdf
- https://blog.paperspace.com/dimension-reduction-with-lle/
Auto-Encoder is a neural network model that compresses the original data and then encodes the compressed information such as
Now suppose we have only unlabeled training examples set
An auto-encoder neural network is an unsupervised learning algorithm
that applies back-propagation, setting the target values to be equal to the inputs. I.e., it uses
Recall that
And we will minimize the objective function
via backpropagation, where
If we want to compress the information of the data set, we only need output of hidden units
- https://en.wikipedia.org/wiki/Autoencoder
- https://www.cs.toronto.edu/~hinton/science.pdf
- https://web.stanford.edu/class/cs294a/sparseAutoencoder.pdf
- http://ufldl.stanford.edu/wiki/index.php/Stacked_Autoencoders
- https://www.elen.ucl.ac.be/Proceedings/esann/esannpdf/es2016-70.pdf
The basic idea of t-SNE is that the similarity should preserve after dimension reduction.
It maps the data
Stochastic Neighbor Embedding (SNE) starts by converting the high-dimensional Euclidean distances between data points into conditional probabilities
that represent similarities. The similarity of data point
Because we are only interested in modeling pairwise similarities, we set the value of
SNE minimizes the sum of cross entropy over all data points using a gradient descent
method. The cost function
In the high-dimensional space, we convert distances into probabilities using a Gaussian distribution.
In the low-dimensional map, we can use a probability distribution that has much heavier tails than a Gaussian to convert distances into probabilities.
In t-SNE, we employ a Student t-distribution with one degree of freedom (which is the same
as a Cauchy distribution) as the heavy-tailed distribution in the low-dimensional map. Using this
distribution, the joint probabilities
- https://lvdmaaten.github.io/tsne/
- https://blog.paperspace.com/dimension-reduction-with-t-sne/
- https://www.analyticsvidhya.com/blog/2017/01/t-sne-implementation-r-python/
- https://distill.pub/2016/misread-tsne
- https://www.wikiwand.com/en/T-distributed_stochastic_neighbor_embedding
Independent component analysis is to find the latent independent factors that make up the observation, i.e, $$ x_j = b_{j1}s_1 + b_{j2}s_2 + \dots + b_{jn} s_n,\forall j\in{1,2,\dots,n} \ x=Bs $$
where
In the ICA model, we assume that each mixture
Different from the principal components in PCA, the independent components are not required to be perpendicular to each other.
The starting point for ICA is the very simple assumption that the components
In order to solve
Intuitively speaking, the key to estimating the ICA model is nongaussianity. Actually, without nongaussianity the
estimation is not possible at all.
The classical measure of nongaussianity is kurtosis or the fourth-order cumulant.
The kurtosis of random variable nongaussianity is given by negentropy
It is proved the surprising result that the principle of network entropy maximization, or “infomax”, is equivalent to maximum likelihood estimation.
ICA is very closely related to the method called blind source separation (BSS) or blind signal separation and see more on Blind Identification of Potentially Underdetermined Mixtures.
- https://www.cs.helsinki.fi/u/ahyvarin/papers/NN00new.pdf
- https://research.ics.aalto.fi/ica/
- http://compneurosci.com/wiki/images/4/42/Intro_to_PCA_and_ICA.pdf
- https://www.wikiwand.com/en/Independent_component_analysis
- http://www.gipsa-lab.grenoble-inp.fr/~pierre.comon/FichiersPdf/polyD16-2006.pdf
- http://arnauddelorme.com/ica_for_dummies/
- https://sccn.ucsd.edu/wiki/Chapter_09:_Decomposing_Data_Using_ICA
- http://cs229.stanford.edu/notes/cs229-notes11.pdf
- Diving Deeper into Dimension Reduction with Independent Components Analysis (ICA)
- https://www.zhihu.com/question/28845451/answer/42292804
- http://www.gipsa-lab.grenoble-inp.fr/~pierre.comon/publications_en.html#book
- https://www.stat.pitt.edu/sungkyu/course/2221Fall13/lec6_FA_ICA.pdf
- https://perso.univ-rennes1.fr/laurent.albera/alberasiteweb/pdf/Albe03-PHD.pdf
- http://fourier.eng.hmc.edu/e161/lectures/ica/index.html
- http://cis.legacy.ics.tkk.fi/aapo/papers/IJCNN99_tutorialweb/
- https://www.cs.helsinki.fi/u/ahyvarin/whatisica.shtml
Projection pursuit is a classical exploratory data analysis method to detect interesting low-dimensional structures in multivariate data. Originally, projection pursuit was applied mostly to data of moderately low dimension. Peter J Huber said in Projection Pursuit:
The most exciting features of projection pursuit is that it is one of few multivariate methods able to bypass the "curse of dimensionality" caused by the fact that high-dimensional space is mostly empty.
PP methods have one serious drawback: their high demand on computer time.
Discussion by Friedman put that
Projection pursuit methods are by no means the only ones to be originally ignored for lack of theoretical justification. Factor analysis, clustering, multidimensional scaling, recursive partitioning, correspondence analysis, soft modeling (partial-least-squares), represent methods that were in common sense for many years before their theoretical underpinnings were well understood. Again, the principal justification for their use was that they made sense heuristically and seemed to work well in a wide variety of situations.
It reduces the data of dimension
The minimal ingredients of an EPP algorithm are then as follows:
- (1) choose a subspace of the desired dimension and project the data onto the subspace,
- (2) compute some index of 'information content' for the projection,
- and (3) iterate 1 and 2 until the index is maximized.
In more mathematical language, EPP involves choosing some
dimension
Assume that the
Friedman and Tukey (1974) proposed to investigate the high-dimensional distribution of
and
The projection pursuit methods can extend to density estimation and regression.
- https://projecteuclid.org/euclid.aos/1176349519
- https://projecteuclid.org/euclid.aos/1176349520
- https://projecteuclid.org/euclid.aos/1176349535
- guynason, Professor of Statistics, University of Bristol
- http://sci-hub.fun/10.1002/wics.23
- Werner Stuetzle, Department of Statistics, University of Washington
- ICA and Projection Pursuit
- https://www.pnas.org/content/115/37/9151
- Projection pursuit in high dimensions
- https://github.com/pavelkomarov/projection-pursuit
- Interactive Projection Pursuit (IPP) by Jiayang Sun, Jeremy Fleischer, Catherine Loader
- Exploratory Projection Pursuit
- A Projection Pursuit framework for supervised dimension reduction of high dimensional small sample datasets
One source of ICA is "general infomax learning principle" well known in machine learning or signal processing community.
However, we can not explain all efficient methods in mathematics or statistics then write it in the textbook.
Self organizing map is not well-known. These networks are based
on competitive learning; the output neurons of the network compete among themselves to
be activated or fired, with the result that only one output neuron, or one neuron per group.
Each node has a specific topological position (an x, y coordinate in the lattice) and contains a vector of weights of the same dimension as the input vectors.
That is to say, if the training data consists of vectors,
Then each node will contain a corresponding weight vector
Training occurs in several steps and over many iterations:
- Each node's weights are initialized.
- A vector is chosen at random from the set of training data and presented to the lattice.
- Every node is examined to calculate which one's weights are most like the input vector.
The winning node is commonly known as the
Best Matching Unit (BMU). - The radius of the neighbourhood of the BMU is now calculated. This is a value that starts large, typically set to the 'radius' of the lattice,
but diminishes each time-step. Any nodes found within this radius are deemed to be inside the BMU's neighbourhood. - Each neighbouring node's (the nodes found in step 4) weights are adjusted to make them more like the input vector. The closer a node is to the BMU, the more its weights get altered.
- Repeat step 2 for N iterations.
- Kohonen Network - Background Information
- https://users.ics.aalto.fi/teuvo/
- http://www.ai-junkie.com/ann/som/som1.html
- http://www.mlab.uiah.fi/~timo/som/thesis-som.html
- Self-Organizing Maps
Diffusion map uses the eigen-vectors to define coordinate system.
https://www.wikiwand.com/en/Diffusion_map
- https://www.wikiwand.com/en/Nonlinear_dimensionality_reduction
- destiny:An R package for diffusion maps, with additional features for large-scale and single cell data
- A short introduction to Diffusion Maps
- pydiffmap: an open-source project to develop a robust and accessible diffusion map code for public use.
- MAT 585: Diffusion Maps by Amit Singer
- http://www.math.pku.edu.cn/teachers/yaoy/Spring2011/
- https://manifoldlearningjl.readthedocs.io/en/latest/ltsa.html
- https://www.cs.cmu.edu/~bapoczos/Classes/ML10715_2015Fall/manifold.html
- http://www.math.pku.edu.cn/teachers/yaoy/Spring2011/lecture11_3_mani.pdf
Uniform Manifold Approximation and Projection (UMAP) is a dimension reduction technique that can be used for visualisation similarly to t-SNE, but also for general non-linear dimension reduction. The algorithm is founded on three assumptions about the data
- The data is uniformly distributed on Riemannian manifold;
- The Riemannian metric is locally constant (or can be approximated as such);
- The manifold is locally connected.
- https://github.com/lmcinnes/umap
- https://umap-learn.readthedocs.io/en/latest/
- https://arxiv.org/abs/1802.03426
In Description Of Intrinsic Dimension 2019, Yoav Freund pointed out that:
It is often the case that very high dimensional data, such as images, can be compressed into low dimensional vectors with small reconstruction error. The dimension of these vectors is the
intrinsic dimensionof the data. We will discuss several techniques for estimating intrinsic dimension and for mapping data vectors to their low-dimensional representations. The ultimate goal is to find streaming algorithms can can process very large dataset in linear or sub-linear time.
In mathematical terms, let the original data be given by the finite sequence of points
Yet, numerous reduction methods do not compute a target embedding dimension but rather rely on an external input parameter. Consequently, the estimation of the intrinsic dimension of a given dataset is essential for the proper functioning of those methods.
- Intrinsic dimension of data representations in deep neural networks
- Intrinsic Dimension Estimation using Simplex Volumes
- Intrinsic dimension estimation for locally undersampled data
- Structures in High-Dimensional Data: Intrinsic Dimension and Cluster Analysis
- Intrinsic Dimension 2019:: Papers
- http://hal.cse.msu.edu/assets/pdfs/papers/2019-cvpr-intrinsic-dimensionality.pdf
- https://people.eng.unimelb.edu.au/baileyj/papers/PID5041135-2.pdf
- Measuring the Intrinsic Dimension of Objective Landscapes
- https://en.wikipedia.org/wiki/Intrinsic_dimension
- https://www.hcm.uni-bonn.de/research/research-areas/ra-b2/
Methods for identifying the dimension:
- Haussdorff dimension, Doubling dimension, epsilon-cover
Lebesgue covering dimension, also called topological dimension or just covering dimension, is defined with respect to a given topological space
Set doubling dimension
One popular example is the relationship of volumes between the unit hypercube and its inscribed d-ball. It might seem surprising at first that the volume of the corresponding d-ball tends to zero very quickly.
The goal is to capture similarity between embeddings, such that the projected distance of similar items in the embedding space is smaller than the dissimilar items. Compared to the standard distance metric learning, it uses deep neural networks to learn a nonlinear mapping to the embedding space. It helps with extreme classification settings with huge number classes, not many examples per class.
- Left and right legs of the network have identical structures (siamese);
- Weights are shared between the siamese networks during training;
- Networks are optimized with a loss function, such as contrastive loss.