\n",
- " \n",
- "## Checkpoint 3\n",
- "\n",
- "Congratulations! You have trained several image translation models now!\n",
- "Please document hyperparameters, snapshots of predictions on validation set, and loss curves for your models and add the final perforance in [this google doc](https://docs.google.com/document/d/1hZWSVRvt9KJEdYu7ib-vFBqAVQRYL8cWaP_vFznu7D8/edit#heading=h.n5u485pmzv2z). We'll discuss our combined results as a group.\n",
- "
"
- ]
- }
- ],
- "metadata": {
- "jupytext": {
- "cell_metadata_filter": "all",
- "main_language": "python"
- }
- },
- "nbformat": 4,
- "nbformat_minor": 5
-}
diff --git a/part_2/README.md b/part_2/README.md
new file mode 100644
index 0000000..6633857
--- /dev/null
+++ b/part_2/README.md
@@ -0,0 +1,85 @@
+# Exercise 6: Image translation - Part 2
+
+This demo script was developed for the DL@MBL 2024 course by Samuel Tonks, Krull Lab University of Birmingham UK, with many inputs and bugfixes from [Eduardo Hirata-Miyasaki](https://github.com/edyoshikun), [Ziwen Liu](https://github.com/ziw-liu) and [Shalin Mehta](https://github.com/mattersoflight) of CZ Biohub San Francisco.
+
+## Image translation (Virtual Staining) via Generative Modelling
+
+In this part of the exercise, we will tackle the same supervised image-to-image translation task but use an alternative approach. Here we will explore a generative modelling approach, specifically a conditional Generative Adversarial Network (cGAN).
+\n",
+ "\n",
+ "## A heads up of what to expect from the training...\n",
+ "
\n",
+ "**- Visualise results**: We can observe how the performance improves over time using the images tab and the sliding window.\n",
+ "
\n",
+ "**- Discriminator Predicted Probabilities**: We plot the discriminator's predicted probabilities that the phase with fluorescence is phase and fluorescence and that the phase with virtual stain is phase with virtual stain. It is typically trained until the discriminator can no longer classify whether or not the generated images are real or fake better than a random guess (p(0.5)). We plot this for both the training and validation datasets.\n",
+ "
\n",
+ "**- Adversarial Loss**: We can formulate the adversarial loss as a Least Squared Error Loss in which for real data the discriminator should output a value close to 1 and for fake data a value close to 0. The generator's goal is to make the discriminator output a value as close to 1 for fake data. We plot the least squared error loss.\n",
+ "
\n",
+ "**- Feature Matching Loss**: Both networks are also trained using the generator feature matching loss which encourages the generator to produce images that contain similar statistics to the real images at each scale. We also plot the feature matching L1 loss for the training and validation sets together to observe the performance and how the model is fitting the data.\n",
+ "
\n",
+ "This implementation allows for the turning on/off of the least-square loss term by setting the opt.no_lsgan flag to the model options. As well as the turning off of the feature matching loss term by setting the opt.no_ganFeat_loss flag to the model options. Something you might want to explore in the next section!
\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "f1e7c664",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "\n",
+ " \n",
+ "## Checkpoint 1\n",
+ "\n",
+ "Congratulations! You should now have a better understanding of how to train a Pix2PixHD GAN model for translating from phase to nuclei. You should also have a good understanding of the different loss components of Pix2PixHD GAN and how they are used to train the model.\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "9f60283b",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0,
+ "title": "\n",
+ " \n",
+ "## Checkpoint 2\n",
+ "Congratulations! You should now have a better understanding the different loss components of Pix2PixHD GAN and how they are used to train the model. You should also have a good understanding of the fit of the model during training on the training and validation datasets.\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6c62a3d5",
+ "metadata": {
+ "cell_marker": "\"\"\""
+ },
+ "source": [
+ "# Part 3: Evaluate performance of the virtual staining on unseen data.\n",
+ "--------------------------------------------------\n",
+ "## Evaluate the performance of the model.\n",
+ "We now look at the same metrics of performance of the previous model. We typically evaluate the model performance on a held out test data. \n",
+ "\n",
+ "Steps:\n",
+ "- Define our model parameters for the pre-trained model (these are the same parameters as shown in earlier cells but copied here for clarity).\n",
+ "- Load the test data.\n",
+ "\n",
+ "We will first load the test data using the same format as the training and validation data. We will then use the model to sample a virtual nuclei staining soltuion from the phase image. We will then evaluate the performance of the model using the following metrics:\n",
+ "\n",
+ "Pixel-level metrics:\n",
+ "- [Peak-Signal-to-Noise-Ratio (PSNR)](https://en.wikipedia.org/wiki/Peak_signal-to-noise_ratio).\n",
+ "- [Structural Similarity Index Measure (SSIM)](https://en.wikipedia.org/wiki/Structural_similarity).\n",
+ "- [Pearson Correlation Coefficient (PCC)](https://en.wikipedia.org/wiki/Pearson_correlation_coefficient).\n",
+ "\n",
+ "Instance-level metrics via [Cellpose masks](https://cellpose.org/):\n",
+ "- [Accuracy](https://en.wikipedia.org/wiki/Accuracy_and_precision#In_binary_classification)\n",
+ "- [Jaccard Index](https://en.wikipedia.org/wiki/Jaccard_index)\n",
+ "- [Dice Score](https://en.wikipedia.org/wiki/S%C3%B8rensen%E2%80%93Dice_coefficient)\n",
+ "- [Mean Average Precision](https://en.wikipedia.org/wiki/Evaluation_measures_(information_retrieval)#Mean_average_precision)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d38b033f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "opt = TestOptions().parse(save=False)\n",
+ "\n",
+ "# Define the parameters for the dataset.\n",
+ "opt.dataroot = output_image_folder\n",
+ "opt.data_type = 16 # Data type of the images.\n",
+ "opt.loadSize = 512 # Size of the loaded phase image.\n",
+ "opt.input_nc = 1 # Number of input channels.\n",
+ "opt.output_nc = 1 # Number of output channels.\n",
+ "opt.target = \"nuclei\" # \"nuclei\" or \"cyto\" depending on your choice of target for virtual stain.\n",
+ "opt.resize_or_crop = \"none\" # Scaling and cropping of images at load time [resize_and_crop|crop|scale_width|scale_width_and_crop|none].\n",
+ "opt.batchSize = 1 # Batch size for training\n",
+ "\n",
+ "# Define the model parameters for the pre-trained model.\n",
+ "\n",
+ "# Define the parameters for the Generator.\n",
+ "opt.ngf = 64 # Number of filters in the generator.\n",
+ "opt.n_downsample_global = 4 # Number of downsampling layers in the generator.\n",
+ "opt.n_blocks_global = 9 # Number of residual blocks in the generator.\n",
+ "opt.n_blocks_local = 3 # Number of residual blocks in the generator.\n",
+ "opt.n_local_enhancers = 1 # Number of local enhancers in the generator.\n",
+ "\n",
+ "# Define the parameters for the Discriminators.\n",
+ "opt.num_D = 3 # Number of discriminators.\n",
+ "opt.n_layers_D = 3 # Number of layers in the discriminator.\n",
+ "opt.ndf = 32 # Number of filters in the discriminator.\n",
+ "\n",
+ "# Define general training parameters.\n",
+ "opt.gpu_ids= [0] # GPU ids to use.\n",
+ "opt.norm = \"instance\" # Normalization layer in the generator.\n",
+ "opt.use_dropout = \"\" # Use dropout in the generator (fixed at 0.2).\n",
+ "opt.batchSize = 8 # Batch size.\n",
+ "\n",
+ "# Define loss functions.\n",
+ "opt.no_vgg_loss = \"\" # Turn off VGG loss\n",
+ "opt.no_ganFeat_loss = \"\" # Turn off feature matching loss\n",
+ "opt.no_lsgan = \"\" # Turn off least square loss\n",
+ "\n",
+ "# Additional Inference parameters\n",
+ "opt.name = f\"dlmbl_vsnuclei\"\n",
+ "opt.how_many = 112 # Number of images to generate.\n",
+ "opt.checkpoints_dir = f\"{top_dir}/model_weights/\" # Path to the model checkpoints.\n",
+ "opt.results_dir = f\"{top_dir}/GAN_code/GANs_MI2I/pre_trained/{opt.name}/inference_results/\" # Path to store the results.\n",
+ "opt.which_epoch = \"latest\" # or specify the epoch number \"40\"\n",
+ "opt.phase = \"test\"\n",
+ "\n",
+ "opt.nThreads = 1 # test code only supports nThreads = 1\n",
+ "opt.batchSize = 1 # test code only supports batchSize = 1\n",
+ "opt.serial_batches = True # no shuffle\n",
+ "opt.no_flip = True # no flip\n",
+ "Path(opt.results_dir).mkdir(parents=True, exist_ok=True)\n",
+ "\n",
+ "# Load the test data.\n",
+ "test_data_loader = CreateDataLoader(opt)\n",
+ "test_dataset = test_data_loader.load_data()\n",
+ "visualizer = Visualizer(opt)\n",
+ "print(f\"Total Test Images = {len(test_data_loader)}\")\n",
+ "# Load pre-trained model\n",
+ "model = create_model(opt)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "dde67288",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Generate & save predictions in the results directory.\n",
+ "inference_model(test_dataset, opt, model)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "95db1b7e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Gather results for evaluation\n",
+ "virtual_stain_paths = sorted([i for i in Path(opt.results_dir).glob(\"**/*.tiff\")])\n",
+ "target_stain_paths = sorted([i for i in Path(f\"{output_image_folder}/{translation_task}/test/\").glob(\"**/*.tiff\")])\n",
+ "phase_paths = sorted([i for i in Path(f\"{output_image_folder}/input/test/\").glob(\"**/*.tiff\")])\n",
+ "assert (len(virtual_stain_paths) == len(target_stain_paths) == len(phase_paths)\n",
+ "), f\"Number of images do not match. {len(virtual_stain_paths)},{len(target_stain_paths)} {len(phase_paths)} \"\n",
+ "\n",
+ "# Create arrays to store the images.\n",
+ "virtual_stains = np.zeros((len(virtual_stain_paths), 512, 512))\n",
+ "target_stains = virtual_stains.copy()\n",
+ "phase_images = virtual_stains.copy()\n",
+ "# Load the images and store them in the arrays.\n",
+ "for index, (v_path, t_path, p_path) in tqdm(\n",
+ " enumerate(zip(virtual_stain_paths, target_stain_paths, phase_paths))\n",
+ "):\n",
+ " virtual_stain = imread(v_path)\n",
+ " phase_image = imread(p_path)\n",
+ " target_stain = imread(t_path)\n",
+ " # Append the images to the arrays.\n",
+ " phase_images[index] = phase_image\n",
+ " target_stains[index] = target_stain\n",
+ " virtual_stains[index] = virtual_stain"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d168855d",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0,
+ "tags": []
+ },
+ "source": [
+ "\n",
+ "\n",
+ "### Task 3.1 Visualise the results of the model on the test set.\n",
+ "\n",
+ "Create a matplotlib plot that visalises random samples of the phase images, target stains, and virtual stains.\n",
+ "If you can incorporate the crop function below to zoom in on the images that would be great!\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "915ddf15",
+ "metadata": {
+ "lines_to_next_cell": 1
+ },
+ "outputs": [],
+ "source": [
+ "# Define a function to crop the images so we can zoom in.\n",
+ "def crop(img, crop_size, loc='center'):\n",
+ " \"\"\"\n",
+ " Crop the input image.\n",
+ "\n",
+ " Parameters:\n",
+ " img (ndarray): The image to be cropped.\n",
+ " crop_size (int): The size of the crop.\n",
+ " loc (str): The type of crop to perform. Can be 'center' or 'random'.\n",
+ "\n",
+ " Returns:\n",
+ " ndarray: The cropped image array.\n",
+ " \"\"\"\n",
+ " # Dimension of input array\n",
+ " width, height = img.shape\n",
+ "\n",
+ " # Generate random coordinates for the crop\n",
+ " max_y = height - crop_size\n",
+ " max_x = max_y\n",
+ "\n",
+ " if loc == 'random':\n",
+ " start_y = np.random.randint(0, max_y + 1)\n",
+ " start_x = np.random.randint(0, max_x + 1)\n",
+ " end_y = start_y + crop_size\n",
+ " end_x = start_x + crop_size\n",
+ " elif loc == 'center':\n",
+ " start_x = (width - crop_size) // 2\n",
+ " start_y = (height - crop_size) // 2\n",
+ " end_y = height - start_y\n",
+ " end_x = width - start_x\n",
+ " else:\n",
+ " raise ValueError(f'Unknown crop type {loc}')\n",
+ "\n",
+ " # Crop array using slicing\n",
+ " crop_array = img[start_x:end_x, start_y:end_y]\n",
+ " return crop_array"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "cc247ac9",
+ "metadata": {
+ "lines_to_next_cell": 0,
+ "tags": [
+ "solution"
+ ]
+ },
+ "outputs": [],
+ "source": [
+ "\n",
+ "##########################\n",
+ "######## Solution ########\n",
+ "##########################\n",
+ "\n",
+ "def visualise_results(phase_images, target_stains, virtual_stains, crop_size=None, loc='center'):\n",
+ " \"\"\"\n",
+ " Visualizes the results of image processing by displaying the phase images, target stains, and virtual stains.\n",
+ " Parameters:\n",
+ " - phase_images (np.array): Array of phase images.\n",
+ " - target_stains (np.array): Array of target stains.\n",
+ " - virtual_stains (np.array): Array of virtual stains.\n",
+ " - crop_size (int, optional): Size of the crop. Defaults to None.\n",
+ " - type (str, optional): Type of crop. Defaults to 'center' but can be 'random.\n",
+ " Returns:\n",
+ " None\n",
+ " \"\"\" \n",
+ " fig, axes = plt.subplots(5, 3, figsize=(15, 20))\n",
+ " sample_indices = np.random.choice(len(phase_images), 5)\n",
+ " for index,sample in enumerate(sample_indices):\n",
+ " if crop_size:\n",
+ " phase_image = crop(phase_images[index], crop_size, loc)\n",
+ " target_stain = crop(target_stains[index], crop_size, loc)\n",
+ " virtual_stain = crop(virtual_stains[index], crop_size, loc)\n",
+ " else:\n",
+ " phase_image = phase_images[index]\n",
+ " target_stain = target_stains[index]\n",
+ " virtual_stain = virtual_stains[index] \n",
+ "\n",
+ " axes[index, 0].imshow(phase_image, cmap=\"gray\")\n",
+ " axes[index, 0].set_title(\"Phase\")\n",
+ " axes[index, 1].imshow(\n",
+ " target_stain,\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stain, 1),\n",
+ " vmax=np.percentile(target_stain, 99),\n",
+ " )\n",
+ " axes[index, 1].set_title(\"Target Fluorescence \")\n",
+ " axes[index, 2].imshow(\n",
+ " virtual_stain,\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stain, 1),\n",
+ " vmax=np.percentile(target_stain, 99),\n",
+ " )\n",
+ " axes[index, 2].set_title(\"Virtual Stain\")\n",
+ " for ax in axes.flatten():\n",
+ " ax.axis(\"off\")\n",
+ " plt.tight_layout()\n",
+ " plt.show()\n",
+ "\n",
+ "visualise_results(phase_images, target_stains,virtual_stains,crop_size=None)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "77b038e6",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0,
+ "tags": []
+ },
+ "source": [
+ "\n",
+ "\n",
+ "### Task 3.2 Compute pixel-level metrics\n",
+ "\n",
+ "Compute the pixel-level metrics for the virtual stains and target stains.\n",
+ "\n",
+ "The following code will compute the following:\n",
+ "- the pixel-based metrics (Pearson correlation, SSIM, PSNR) for the virtual stains and target stains.\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "7ea32c4c",
+ "metadata": {
+ "lines_to_next_cell": 0
+ },
+ "outputs": [],
+ "source": [
+ "\n",
+ "# Define the function to perform minmax normalization which is required for the pixel-level metrics.\n",
+ "def min_max_scale(input):\n",
+ " return (input - np.min(input)) / (np.max(input) - np.min(input))\n",
+ "\n",
+ "# Create a dataframe to store the pixel-level metrics.\n",
+ "test_pixel_metrics = pd.DataFrame(\n",
+ " columns=[\"model\", \"fov\",\"pearson_nuc\", \"ssim_nuc\", \"psnr_nuc\"]\n",
+ ")\n",
+ "\n",
+ "# Compute the pixel-level metrics.\n",
+ "for i, (target_stain, predicted_stain) in tqdm(enumerate(zip(target_stains, virtual_stains))):\n",
+ " fov = str(virtual_stain_paths[i]).split(\"/\")[-1].split(\".\")[0]\n",
+ " minmax_norm_target = min_max_scale(target_stain)\n",
+ " minmax_norm_predicted = min_max_scale(predicted_stain)\n",
+ " \n",
+ " # Compute SSIM\n",
+ " ssim_nuc = metrics.structural_similarity(\n",
+ " minmax_norm_target, minmax_norm_predicted, data_range=1\n",
+ " )\n",
+ " # Compute Pearson correlation\n",
+ " pearson_nuc = np.corrcoef(\n",
+ " minmax_norm_target.flatten(), minmax_norm_predicted.flatten()\n",
+ " )[0, 1]\n",
+ " # Compute PSNR\n",
+ " psnr_nuc = metrics.peak_signal_noise_ratio(\n",
+ " minmax_norm_target, minmax_norm_predicted, data_range=1\n",
+ " )\n",
+ " \n",
+ " test_pixel_metrics.loc[len(test_pixel_metrics)] = {\n",
+ " \"model\": \"pix2pixHD\",\n",
+ " \"fov\":fov,\n",
+ " \"pearson_nuc\": pearson_nuc,\n",
+ " \"ssim_nuc\": ssim_nuc,\n",
+ " \"psnr_nuc\": psnr_nuc, \n",
+ " }\n",
+ " \n",
+ "test_pixel_metrics.boxplot(\n",
+ " column=[\"pearson_nuc\", \"ssim_nuc\"],\n",
+ " rot=30,\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "e09b3843",
+ "metadata": {
+ "lines_to_next_cell": 0
+ },
+ "outputs": [],
+ "source": [
+ "test_pixel_metrics.boxplot(\n",
+ " column=[\"psnr_nuc\"],\n",
+ " rot=30,\n",
+ ")"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "af1a13e0",
+ "metadata": {
+ "lines_to_next_cell": 0
+ },
+ "outputs": [],
+ "source": [
+ "test_pixel_metrics.head()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "83bbcee9",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "## Inference Pixel-level Results\n",
+ "Please note down your thoughts about the following questions...\n",
+ "\n",
+ "- What do these metrics tells us about the performance of the model?\n",
+ "- How do the pixel-level metrics compare to the regression-based approach?\n",
+ "- Could these metrics be skewed by the presence of hallucinations or background pilxels in the virtual stains?"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "74de240b",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "\n",
+ "\n",
+ "### Task 3.3 Compute instance-level metrics\n",
+ "\n",
+ "- Compute the instance-level metrics for the virtual stains and target stains.\n",
+ "- Instance metrics include the accuracy (average correct predictions with 0.5 threshold), jaccard index (intersection over union (IoU)) dice score (2x intersection over union), mean average precision, mean average precision at 50% IoU, mean average precision at 75% IoU, and mean average recall at 100% IoU.\n",
+ "\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "25bfb934",
+ "metadata": {
+ "lines_to_next_cell": 0
+ },
+ "outputs": [],
+ "source": [
+ "\n",
+ "# Use the same function as previous part to extract the nuclei masks from pre-trained cellpose model.\n",
+ "def cellpose_segmentation(prediction:ArrayLike,target:ArrayLike)->Tuple[torch.ShortTensor]:\n",
+ " # NOTE these are hardcoded for this notebook and A549 dataset\n",
+ " device = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\n",
+ " cp_nuc_kwargs = {\n",
+ " \"diameter\": 65,\n",
+ " \"cellprob_threshold\": 0.0, \n",
+ " }\n",
+ " cellpose_model = models.CellposeModel(\n",
+ " gpu=True, model_type='nuclei', device=torch.device(device)\n",
+ " )\n",
+ " pred_label, _, _ = cellpose_model.eval(prediction, **cp_nuc_kwargs)\n",
+ " target_label, _, _ = cellpose_model.eval(target, **cp_nuc_kwargs)\n",
+ "\n",
+ " pred_label = pred_label.astype(np.int32)\n",
+ " target_label = target_label.astype(np.int32)\n",
+ " pred_label = torch.ShortTensor(pred_label)\n",
+ " target_label = torch.ShortTensor(target_label)\n",
+ "\n",
+ " return (pred_label,target_label)\n",
+ "\n",
+ "# Define dataframe to store the segmentation metrics.\n",
+ "test_segmentation_metrics= pd.DataFrame(\n",
+ " columns=[\"model\", \"fov\",\"masks_per_fov\",\"accuracy\",\"dice\",\"jaccard\",\"mAP\",\"mAP_50\",\"mAP_75\",\"mAR_100\"]\n",
+ ")\n",
+ "# Define tuple to store the segmentation results. Each value in the tuple is a dictionary containing the model name, fov, predicted label, predicted stain, target label, and target stain.\n",
+ "segmentation_results = ()\n",
+ "\n",
+ "for i, (target_stain, predicted_stain) in tqdm(enumerate(zip(target_stains, virtual_stains))):\n",
+ " fov = str(virtual_stain_paths)[i].spilt(\"/\")[-1].split(\".\")[0]\n",
+ " minmax_norm_target = min_max_scale(target_stain)\n",
+ " minmax_norm_predicted = min_max_scale(predicted_stain)\n",
+ " # Compute the segmentation masks.\n",
+ " pred_label, target_label = cellpose_segmentation(minmax_norm_predicted, minmax_norm_target)\n",
+ " # Binary labels\n",
+ " pred_label_binary = pred_label > 0\n",
+ " target_label_binary = target_label > 0\n",
+ "\n",
+ " # Use Coco metrics to get mean average precision\n",
+ " coco_metrics = mean_average_precision(pred_label, target_label)\n",
+ " # Find unique number of labels\n",
+ " num_masks_fov = len(np.unique(pred_label))\n",
+ " # Find unique number of labels\n",
+ " num_masks_fov = len(np.unique(pred_label))\n",
+ " # Compute the segmentation metrics.\n",
+ " test_segmentation_metrics.loc[len(test_segmentation_metrics)] = {\n",
+ " \"model\": \"pix2pixHD\",\n",
+ " \"fov\":fov,\n",
+ " \"masks_per_fov\": num_masks_fov,\n",
+ " \"accuracy\": accuracy(pred_label_binary, target_label_binary, task=\"binary\").item(),\n",
+ " \"dice\": dice(pred_label_binary, target_label_binary).item(),\n",
+ " \"jaccard\": jaccard_index(pred_label_binary, target_label_binary, task=\"binary\").item(),\n",
+ " \"mAP\":coco_metrics[\"map\"].item(),\n",
+ " \"mAP_50\":coco_metrics[\"map_50\"].item(),\n",
+ " \"mAP_75\":coco_metrics[\"map_75\"].item(),\n",
+ " \"mAR_100\":coco_metrics[\"mar_100\"].item()\n",
+ " }\n",
+ " # Store the segmentation results.\n",
+ " segmentation_result = {\n",
+ " \"model\": \"pix2pixHD\",\n",
+ " \"fov\":fov,\n",
+ " \"phase_image\": phase_images[i],\n",
+ " \"pred_label\": pred_label,\n",
+ " \"pred_stain\": predicted_stain,\n",
+ " \"target_label\": target_label,\n",
+ " \"target_stain\": target_stain,\n",
+ " }\n",
+ " segmentation_results += (segmentation_result,)\n",
+ "\n",
+ "test_segmentation_metrics.head()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "3c19de9e",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Define function to visualize the segmentation results.\n",
+ "def visualise_results_and_masks(segmentation_results: Tuple[dict], segmentation_metrics: pd.DataFrame, rows: int = 5, crop_size: int = None, crop_type: str = 'center'):\n",
+ "\n",
+ " # Sample a subset of the segmentation results.\n",
+ " sample_indices = np.random.choice(len(phase_images),rows)\n",
+ " print(sample_indices)\n",
+ " segmentation_metrics = segmentation_metrics.iloc[sample_indices,:]\n",
+ " segmentation_results = [segmentation_results[i] for i in sample_indices]\n",
+ " # Define the figure and axes.\n",
+ " fig, axes = plt.subplots(rows, 5, figsize=(rows*3, 15))\n",
+ "\n",
+ " # Visualize the segmentation results.\n",
+ " for i in range(len((segmentation_results))):\n",
+ " segmentation_metric = segmentation_metrics.iloc[i]\n",
+ " result = segmentation_results[i]\n",
+ " phase_image = result[\"phase_image\"]\n",
+ " target_stain = result[\"target_stain\"]\n",
+ " target_label = result[\"target_label\"]\n",
+ " pred_stain = result[\"pred_stain\"]\n",
+ " pred_label = result[\"pred_label\"]\n",
+ " # Crop the images if required. Zoom into instances\n",
+ " if crop_size is not None:\n",
+ " phase_image = crop(phase_image, crop_size, crop_type)\n",
+ " target_stain = crop(target_stain, crop_size, crop_type)\n",
+ " target_label = crop(target_label, crop_size, crop_type)\n",
+ " pred_stain = crop(pred_stain, crop_size, crop_type)\n",
+ " pred_label = crop(pred_label, crop_size, crop_type)\n",
+ " \n",
+ " axes[i, 0].imshow(phase_image, cmap=\"gray\")\n",
+ " axes[i, 0].set_title(\"Phase\")\n",
+ " axes[i, 1].imshow(\n",
+ " target_stain,\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stain, 1),\n",
+ " vmax=np.percentile(target_stain, 99),\n",
+ " )\n",
+ " axes[i, 1].set_title(\"Target Fluorescence\")\n",
+ " axes[i, 2].imshow(pred_stain, cmap=\"gray\")\n",
+ " axes[i, 2].set_title(\"Virtual Stain\")\n",
+ " axes[i, 3].imshow(target_label, cmap=\"inferno\")\n",
+ " axes[i, 3].set_title(\"Target Fluorescence Mask\")\n",
+ " axes[i, 4].imshow(pred_label, cmap=\"inferno\")\n",
+ " # Add Metric values to the title\n",
+ " axes[i, 4].set_title(f\"Virtual Stain Mask\\nAcc:{segmentation_metric['accuracy']:.2f} Dice:{segmentation_metric['dice']:.2f}\\nJaccard:{segmentation_metric['jaccard']:.2f} MAP:{segmentation_metric['mAP']:.2f}\")\n",
+ " # Turn off the axes.\n",
+ " for ax in axes.flatten():\n",
+ " ax.axis(\"off\")\n",
+ "\n",
+ " plt.tight_layout()\n",
+ " plt.show()\n",
+ " \n",
+ "visualise_results_and_masks(segmentation_results,test_segmentation_metrics, crop_size=256, crop_type='center')"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "6f11e65a",
+ "metadata": {
+ "lines_to_next_cell": 0
+ },
+ "source": []
+ },
+ {
+ "cell_type": "markdown",
+ "id": "488b6367",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "## Inference Instance-level Results\n",
+ "Please note down your thoughts about the following questions...\n",
+ "\n",
+ "- What do these metrics tells us about the performance of the model?\n",
+ "- How does the performance compare to when looking at pixel-level metrics?"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "9395a312",
+ "metadata": {
+ "cell_marker": "\"\"\""
+ },
+ "source": [
+ "\n",
+ " \n",
+ "## Checkpoint 3\n",
+ "\n",
+ "Congratulations! You have generated predictions from a pre-trained model and evaluated the performance of the model on unseen data. You have computed pixel-level metrics and instance-level metrics to evaluate the performance of the model. You may have also began training your own Pix2PixHD GAN models with alternative hyperparameters.\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "81329322",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "# Part 4. Visualise Regression vs Generative Modelling Approaches\n",
+ "--------------------------------------------------"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "6665fe93",
+ "metadata": {
+ "lines_to_next_cell": 0,
+ "tags": [
+ "solution"
+ ]
+ },
+ "outputs": [],
+ "source": [
+ "\n",
+ "##########################\n",
+ "######## Solution ########\n",
+ "##########################\n",
+ "\n",
+ "def visualise_both_methods(\n",
+ " phase_images: np.array, target_stains: np.array, pix2pixHD_results: np.array, viscy_results: np.array,crop_size=None,crop_type='center'\n",
+ "):\n",
+ " fig, axes = plt.subplots(5, 4, figsize=(15, 15))\n",
+ " sample_indices = np.random.choice(len(phase_images), 5)\n",
+ " if crop_size is not None:\n",
+ " phase_image = crop(phase_image, crop_size, crop_type)\n",
+ " target_stain = crop(target_stain, crop_size, crop_type)\n",
+ " target_label = crop(target_label, crop_size, crop_type)\n",
+ " pred_stain = crop(pred_stain, crop_size, crop_type)\n",
+ " pred_label = crop(pred_label, crop_size, crop_type)\n",
+ "\n",
+ " for i, idx in enumerate(sample_indices):\n",
+ " axes[i, 0].imshow(phase_images[idx], cmap=\"gray\")\n",
+ " axes[i, 0].set_title(\"Phase\")\n",
+ " axes[i, 0].axis(\"off\")\n",
+ " axes[i, 1].imshow(\n",
+ " target_stains[idx],\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stains[idx], 1),\n",
+ " vmax=np.percentile(target_stains[idx], 99),\n",
+ " )\n",
+ " axes[i, 1].set_title(\"Nuclei\")\n",
+ " axes[i, 1].axis(\"off\")\n",
+ " \n",
+ " axes[i, 2].imshow(\n",
+ " viscy_results[idx],\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stains[idx], 1),\n",
+ " vmax=np.percentile(target_stains[idx], 99),\n",
+ " )\n",
+ " axes[i, 2].set_title(\"Regression\\nVirtual Stain\")\n",
+ " axes[i, 2].axis(\"off\")\n",
+ " \n",
+ " axes[i, 3].imshow(\n",
+ " pix2pixHD_results[idx],\n",
+ " cmap=\"gray\",\n",
+ " vmin=np.percentile(target_stains[idx], 1),\n",
+ " vmax=np.percentile(target_stains[idx], 99),\n",
+ " )\n",
+ " axes[i, 3].set_title(\"Pix2PixHD GAN\\nVirtual Stain\")\n",
+ " axes[i, 3].axis(\"off\")\n",
+ " plt.tight_layout()\n",
+ " plt.show()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "35f29396",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "\n",
+ " \n",
+ "## Checkpoint 4\n",
+ "\n",
+ "Congratulations! You should now have a better understanding of the difference in performance for image translation when approaching the problem using a regression vs. generative modelling approaches!\n",
+ "\n",
+ "
"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "ec2331fd",
+ "metadata": {
+ "cell_marker": "\"\"\"",
+ "lines_to_next_cell": 0
+ },
+ "source": [
+ "# Part 5: BONUS: Sample different virtual staining solutions from the GAN using MC-Dropout and explore the uncertainty in the virtual stain predictions.\n",
+ "--------------------------------------------------\n",
+ "Steps:\n",
+ "- Load the pre-trained model.\n",
+ "- Generate multiple predictions for the same input image.\n",
+ "- Compute the pixel-wise variance across the predictions.\n",
+ "- Visualise the pixel-wise variance to explore the uncertainty in the virtual stain predictions."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "1ea346ff",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Use the same model and dataloaders as before.\n",
+ "# Load the test data.\n",
+ "test_data_loader = CreateDataLoader(opt)\n",
+ "test_dataset = test_data_loader.load_data()\n",
+ "visualizer = Visualizer(opt)\n",
+ "\n",
+ "# Load pre-trained model\n",
+ "opt.variational_inf_runs = 100 # Number of samples per phase input\n",
+ "opt.variation_inf_path = f\"./GAN_code/GANs_MI2I/pre_trained/{opt.name}/samples/\" # Path to store the samples.\n",
+ "opt.results_dir = f\"{top_dir}/GAN_code/GANs_MI2I/pre_trained/dlmbl_vsnuclei/sampling_results\"\n",
+ "opt.dropout_variation_inf = True # Use dropout during inference.\n",
+ "model = create_model(opt)\n",
+ "# Generate & save predictions in the variation_inf_path directory.\n",
+ "sampling(test_dataset, opt, model)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "d359ea46",
+ "metadata": {
+ "lines_to_next_cell": 1
+ },
+ "outputs": [],
+ "source": [
+ "# Visualise Samples \n",
+ "samples = sorted([i for i in Path(f\"{opt.results_dir}\").glob(\"**/*.tif*\")])\n",
+ "assert len(samples) == 5\n",
+ "# Create arrays to store the images.\n",
+ "sample_images = np.zeros((5,100, 512, 512)) # (samples, images, height, width)\n",
+ "# Load the images and store them in the arrays.\n",
+ "for index, sample_path in tqdm(enumerate(samples)):\n",
+ " sample_image = imread(sample_path)\n",
+ " # Append the images to the arrays.\n",
+ " sample_images[index] = sample_image"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": null,
+ "id": "26c430fc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Create a matplotlib plot with animation through images.\n",
+ "def animate_images(images):\n",
+ " # Expecting images to have shape (frames, height, width)\n",
+ " fig, ax = plt.subplots()\n",
+ " ax.axis('off')\n",
+ " \n",
+ " # Make sure images are in (frames, height, width) order\n",
+ " images = images.transpose(0, 2, 1) if images.shape[1] == images.shape[2] else images\n",
+ " \n",
+ " imgs = []\n",
+ " for i in range(min(100, len(images))): # Ensure you don't exceed the number of frames\n",
+ " im = ax.imshow(images[i], animated=True)\n",
+ " imgs.append([im])\n",
+ "\n",
+ " ani = animation.ArtistAnimation(fig, imgs, interval=100, blit=False, repeat_delay=1000)\n",
+ " \n",
+ " # Display the animation\n",
+ " # plt.close(fig)\n",
+ " display(HTML(ani.to_jshtml()))\n",
+ "\n",
+ "# Example call with sample_images[0]\n",
+ "animate_images(sample_images[0])"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "c2e73539",
+ "metadata": {
+ "cell_marker": "\"\"\""
+ },
+ "source": [
+ "\n",
+ " \n",
+ "## Checkpoint 5\n",
+ "\n",
+ "Congratulations! This is the end of the conditional generative modelling approach to image translation notebook. You have trained and examined the loss components of Pix2PixHD GAN. You have compared the results of a regression-based approach vs. generative modelling approach and explored the variability in virtual staining solutions. I hope you have enjoyed learning experience!\n",
+ "
"
+ ]
+ }
+ ],
+ "metadata": {
+ "jupytext": {
+ "cell_metadata_filter": "all",
+ "main_language": "python"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.8.19"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
diff --git a/part_2/solution.py b/part_2/solution.py
new file mode 100644
index 0000000..498b9f6
--- /dev/null
+++ b/part_2/solution.py
@@ -0,0 +1,927 @@
+# %% [markdown]
+"""
+# A Generative Modelling Approach to Image translation
+Written by [Samuel Tonks](https://github.com/Tonks684), Krull Lab University of Birmingham UK, with many inputs and bugfixes from [Eduardo Hirata-Miyasaki](https://github.com/edyoshikun), [Ziwen Liu](https://github.com/ziw-liu) and [Shalin Mehta](https://github.com/mattersoflight) of CZ Biohub San Francisco.
+"""
+
+# %% [markdown]
+"""
+## Overview
+
+In part 2 of the image_translation exercise, we will predict fluorescence images of nuclei markers only from quantitative phase images of cells, using a specific type of generative model called a Conditional Generative Adversarial Network (cGAN). In contrast to a regression-based approach, cGANs learn to map from the phase contrast domain to a distirbution of virtual staining solutions. In this work we will utilise the [Pix2PixHD GAN](https://arxiv.org/abs/1711.11585) used in our recent [virtual staining works](https://ieeexplore.ieee.org/abstract/document/10230501?casa_token=NEyrUDqvFfIAAAAA:tklGisf9BEKWVjoZ6pgryKvLbF6JyurOu5Jrgoia1QQLpAMdCSlP9gMa02f3w37PvVjdiWCvFhA). For more details on the architecture and loss components of cGANs and Pix2PixHD GAN please see the READ.me.
+
+During this exercise will assess the different loss components of a pre-trained Pix2PixHD for the virtual nuclei staining task. We will then evaluate the performance of the model on unseen data using the same pixel-level and instance-level metrics as in the previous section. We will compare the performance of the Pix2PixHD GAN with the regression-based model Viscy. Finally, as a bonus, we will explore the variability and uncertainty in the virtual stain predictions using [MC-Dropout](https://arxiv.org/abs/1506.02142).
+
+## References
+- [Wang, T. et al. (2018) High-resolution image synthesis and semantic manipulation with conditional GANs](https://arxiv.org/abs/1711.11585)
+- [Tonks, S. et al. (2023) Evaluating virtual staining for high-throughput screening](https://ieeexplore.ieee.org/abstract/document/10230501?casa_token=NEyrUDqvFfIAAAAA:tklGisf9BEKWVjoZ6pgryKvLbF6JyurOu5Jrgoia1QQLpAMdCSlP9gMa02f3w37PvVjdiWCvFhA)
+- [Gal, Y. et al. (2016) Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning](https://arxiv.org/abs/1506.02142)
+
+"""
+# %% [markdown]
+"""
+## Goals
+
+This part of the exercise is organized in 5 parts.
+
+## Checkpoint 1
+
+Congratulations! You should now have a better understanding of how to train a Pix2PixHD GAN model for translating from phase to nuclei. You should also have a good understanding of the different loss components of Pix2PixHD GAN and how they are used to train the model.
+
+
+"""
+# %%
+=======
+# %% [markdown]
+#
+## Training Results
+#Please note down your thoughts about the following questions...
+#
+# **- What do you notice about the virtual staining predictions? How do they appear compared to the regression-based approach? Can you spot any hallucinations?**
+# **- What do you notice about the probabilities of the discriminators? How do the values compare during training compared to validation?**
+# **- What do you notice about the feature matching L1 loss?**
+# **- What do you notice about the least-square loss?**
+#
+
+# %% [markdown]
+#
+#
+#
+>>>>>>> ff0946289bce4234aedb07af642c943d6d40dd24
+## Checkpoint 2
+#Congratulations! You should now have a better understanding the different loss components of Pix2PixHD GAN and how they are used to train the model. You should also have a good understanding of the fit of the model during training on the training and validation datasets.
+#
+#
+
+
+# %% [markdown]
+"""
+# Part 3: Evaluate performance of the virtual staining on unseen data.
+--------------------------------------------------
+## Evaluate the performance of the model.
+We now look at the same metrics of performance of the previous model. We typically evaluate the model performance on a held out test data.
+
+Steps:
+- Define our model parameters for the pre-trained model (these are the same parameters as shown in earlier cells but copied here for clarity).
+- Load the test data.
+
+We will first load the test data using the same format as the training and validation data. We will then use the model to sample a virtual nuclei staining soltuion from the phase image. We will then evaluate the performance of the model using the following metrics:
+
+Pixel-level metrics:
+- [Peak-Signal-to-Noise-Ratio (PSNR)](https://en.wikipedia.org/wiki/Peak_signal-to-noise_ratio).
+- [Structural Similarity Index Measure (SSIM)](https://en.wikipedia.org/wiki/Structural_similarity).
+- [Pearson Correlation Coefficient (PCC)](https://en.wikipedia.org/wiki/Pearson_correlation_coefficient).
+
+Instance-level metrics via [Cellpose masks](https://cellpose.org/):
+- [Accuracy](https://en.wikipedia.org/wiki/Accuracy_and_precision#In_binary_classification)
+- [Jaccard Index](https://en.wikipedia.org/wiki/Jaccard_index)
+- [Dice Score](https://en.wikipedia.org/wiki/S%C3%B8rensen%E2%80%93Dice_coefficient)
+- [Mean Average Precision](https://en.wikipedia.org/wiki/Evaluation_measures_(information_retrieval)#Mean_average_precision)
+"""
+
+# %%
+opt = TestOptions().parse(save=False)
+
+# Define the parameters for the dataset.
+opt.dataroot = output_image_folder
+opt.data_type = 16 # Data type of the images.
+opt.loadSize = 512 # Size of the loaded phase image.
+opt.input_nc = 1 # Number of input channels.
+opt.output_nc = 1 # Number of output channels.
+opt.target = "nuclei" # "nuclei" or "cyto" depending on your choice of target for virtual stain.
+opt.resize_or_crop = "none" # Scaling and cropping of images at load time [resize_and_crop|crop|scale_width|scale_width_and_crop|none].
+opt.batchSize = 1 # Batch size for training
+
+# Define the model parameters for the pre-trained model.
+
+# Define the parameters for the Generator.
+opt.ngf = 64 # Number of filters in the generator.
+opt.n_downsample_global = 4 # Number of downsampling layers in the generator.
+opt.n_blocks_global = 9 # Number of residual blocks in the generator.
+opt.n_blocks_local = 3 # Number of residual blocks in the generator.
+opt.n_local_enhancers = 1 # Number of local enhancers in the generator.
+
+# Define the parameters for the Discriminators.
+opt.num_D = 3 # Number of discriminators.
+opt.n_layers_D = 3 # Number of layers in the discriminator.
+opt.ndf = 32 # Number of filters in the discriminator.
+
+# Define general training parameters.
+opt.gpu_ids= [0] # GPU ids to use.
+opt.norm = "instance" # Normalization layer in the generator.
+opt.use_dropout = "" # Use dropout in the generator (fixed at 0.2).
+opt.batchSize = 8 # Batch size.
+
+# Define loss functions.
+opt.no_vgg_loss = "" # Turn off VGG loss
+opt.no_ganFeat_loss = "" # Turn off feature matching loss
+opt.no_lsgan = "" # Turn off least square loss
+
+# Additional Inference parameters
+opt.name = f"dlmbl_vsnuclei"
+opt.how_many = 112 # Number of images to generate.
+opt.checkpoints_dir = f"{top_dir}/model_weights/" # Path to the model checkpoints.
+opt.results_dir = f"{top_dir}/GAN_code/GANs_MI2I/pre_trained/{opt.name}/inference_results/" # Path to store the results.
+opt.which_epoch = "latest" # or specify the epoch number "40"
+opt.phase = "test"
+
+opt.nThreads = 1 # test code only supports nThreads = 1
+opt.batchSize = 1 # test code only supports batchSize = 1
+opt.serial_batches = True # no shuffle
+opt.no_flip = True # no flip
+Path(opt.results_dir).mkdir(parents=True, exist_ok=True)
+
+# Load the test data.
+test_data_loader = CreateDataLoader(opt)
+test_dataset = test_data_loader.load_data()
+visualizer = Visualizer(opt)
+print(f"Total Test Images = {len(test_data_loader)}")
+# Load pre-trained model
+model = create_model(opt)
+
+# %%
+# Generate & save predictions in the results directory.
+inference_model(test_dataset, opt, model)
+
+# %%
+# Gather results for evaluation
+virtual_stain_paths = sorted([i for i in Path(opt.results_dir).glob("**/*.tiff")])
+target_stain_paths = sorted([i for i in Path(f"{output_image_folder}/{translation_task}/test/").glob("**/*.tiff")])
+phase_paths = sorted([i for i in Path(f"{output_image_folder}/input/test/").glob("**/*.tiff")])
+assert (len(virtual_stain_paths) == len(target_stain_paths) == len(phase_paths)
+), f"Number of images do not match. {len(virtual_stain_paths)},{len(target_stain_paths)} {len(phase_paths)} "
+
+# Create arrays to store the images.
+virtual_stains = np.zeros((len(virtual_stain_paths), 512, 512))
+target_stains = virtual_stains.copy()
+phase_images = virtual_stains.copy()
+# Load the images and store them in the arrays.
+for index, (v_path, t_path, p_path) in tqdm(
+ enumerate(zip(virtual_stain_paths, target_stain_paths, phase_paths))
+):
+ virtual_stain = imread(v_path)
+ phase_image = imread(p_path)
+ target_stain = imread(t_path)
+ # Append the images to the arrays.
+ phase_images[index] = phase_image
+ target_stains[index] = target_stain
+ virtual_stains[index] = virtual_stain
+
+# %% [markdown] tags=[]
+"""
+
+
+### Task 3.1 Visualise the results of the model on the test set.
+
+Create a matplotlib plot that visalises random samples of the phase images, target stains, and virtual stains.
+If you can incorporate the crop function below to zoom in on the images that would be great!
+
+"""
+# %%
+# Define a function to crop the images so we can zoom in.
+def crop(img, crop_size, loc='center'):
+ """
+ Crop the input image.
+
+ Parameters:
+ img (ndarray): The image to be cropped.
+ crop_size (int): The size of the crop.
+ loc (str): The type of crop to perform. Can be 'center' or 'random'.
+
+ Returns:
+ ndarray: The cropped image array.
+ """
+ # Dimension of input array
+ width, height = img.shape
+
+ # Generate random coordinates for the crop
+ max_y = height - crop_size
+ max_x = max_y
+
+ if loc == 'random':
+ start_y = np.random.randint(0, max_y + 1)
+ start_x = np.random.randint(0, max_x + 1)
+ end_y = start_y + crop_size
+ end_x = start_x + crop_size
+ elif loc == 'center':
+ start_x = (width - crop_size) // 2
+ start_y = (height - crop_size) // 2
+ end_y = height - start_y
+ end_x = width - start_x
+ else:
+ raise ValueError(f'Unknown crop type {loc}')
+
+ # Crop array using slicing
+ crop_array = img[start_x:end_x, start_y:end_y]
+ return crop_array
+
+# %% tags=["task"]
+##########################
+######## TODO ########
+##########################
+
+def visualise_results():
+ # Your code here
+ pass
+
+
+# %% tags=["solution"]
+
+##########################
+######## Solution ########
+##########################
+
+def visualise_results(phase_images, target_stains, virtual_stains, crop_size=None, loc='center'):
+ """
+ Visualizes the results of image processing by displaying the phase images, target stains, and virtual stains.
+ Parameters:
+ - phase_images (np.array): Array of phase images.
+ - target_stains (np.array): Array of target stains.
+ - virtual_stains (np.array): Array of virtual stains.
+ - crop_size (int, optional): Size of the crop. Defaults to None.
+ - type (str, optional): Type of crop. Defaults to 'center' but can be 'random.
+ Returns:
+ None
+ """
+ fig, axes = plt.subplots(5, 3, figsize=(15, 20))
+ sample_indices = np.random.choice(len(phase_images), 5)
+ for index,sample in enumerate(sample_indices):
+ if crop_size:
+ phase_image = crop(phase_images[index], crop_size, loc)
+ target_stain = crop(target_stains[index], crop_size, loc)
+ virtual_stain = crop(virtual_stains[index], crop_size, loc)
+ else:
+ phase_image = phase_images[index]
+ target_stain = target_stains[index]
+ virtual_stain = virtual_stains[index]
+
+ axes[index, 0].imshow(phase_image, cmap="gray")
+ axes[index, 0].set_title("Phase")
+ axes[index, 1].imshow(
+ target_stain,
+ cmap="gray",
+ vmin=np.percentile(target_stain, 1),
+ vmax=np.percentile(target_stain, 99),
+ )
+ axes[index, 1].set_title("Target Fluorescence ")
+ axes[index, 2].imshow(
+ virtual_stain,
+ cmap="gray",
+ vmin=np.percentile(virtual_stain, 1),
+ vmax=np.percentile(virtual_stain, 99),
+ )
+ axes[index, 2].set_title("Virtual Stain")
+ for ax in axes.flatten():
+ ax.axis("off")
+ plt.tight_layout()
+ plt.show()
+
+visualise_results(phase_images, target_stains,virtual_stains,crop_size=None)
+
+# %% [markdown] tags=[]
+"""
+
+
+### Task 3.2 Compute pixel-level metrics
+
+Compute the pixel-level metrics for the virtual stains and target stains.
+
+The following code will compute the following:
+- the pixel-based metrics (Pearson correlation, SSIM, PSNR) for the virtual stains and target stains.
+
+
+"""
+# %%
+
+# Define the function to perform minmax normalization which is required for the pixel-level metrics.
+def min_max_scale(input):
+ return (input - np.min(input)) / (np.max(input) - np.min(input))
+
+# Create a dataframe to store the pixel-level metrics.
+test_pixel_metrics = pd.DataFrame(
+ columns=["model", "fov","pearson_nuc", "ssim_nuc", "psnr_nuc"]
+)
+
+# Compute the pixel-level metrics.
+for i, (target_stain, predicted_stain) in tqdm(enumerate(zip(target_stains, virtual_stains))):
+ fov = str(virtual_stain_paths[i]).split("/")[-1].split(".")[0]
+ minmax_norm_target = min_max_scale(target_stain)
+ minmax_norm_predicted = min_max_scale(predicted_stain)
+
+ # Compute SSIM
+ ssim_nuc = metrics.structural_similarity(
+ minmax_norm_target, minmax_norm_predicted, data_range=1
+ )
+ # Compute Pearson correlation
+ pearson_nuc = np.corrcoef(
+ minmax_norm_target.flatten(), minmax_norm_predicted.flatten()
+ )[0, 1]
+ # Compute PSNR
+ psnr_nuc = metrics.peak_signal_noise_ratio(
+ minmax_norm_target, minmax_norm_predicted, data_range=1
+ )
+
+ test_pixel_metrics.loc[len(test_pixel_metrics)] = {
+ "model": "pix2pixHD",
+ "fov":fov,
+ "pearson_nuc": pearson_nuc,
+ "ssim_nuc": ssim_nuc,
+ "psnr_nuc": psnr_nuc,
+ }
+
+test_pixel_metrics.boxplot(
+ column=["pearson_nuc", "ssim_nuc"],
+ rot=30,
+)
+# %%
+test_pixel_metrics.boxplot(
+ column=["psnr_nuc"],
+ rot=30,
+)
+# %%
+test_pixel_metrics.head()
+# %% [markdown]
+"""
+## Inference Pixel-level Results
+Please note down your thoughts about the following questions...
+
+- What do these metrics tells us about the performance of the model?
+- How do the pixel-level metrics compare to the regression-based approach?
+- Could these metrics be skewed by the presence of hallucinations or background pilxels in the virtual stains?
+
+"""
+# %% [markdown]
+
+"""
+
+
+### Task 3.3 Compute instance-level metrics
+
+- Compute the instance-level metrics for the virtual stains and target stains.
+- Instance metrics include the accuracy (average correct predictions with 0.5 threshold), jaccard index (intersection over union (IoU)) dice score (2x intersection over union), mean average precision, mean average precision at 50% IoU, mean average precision at 75% IoU, and mean average recall at 100% IoU.
+
+
+
+"""
+# %%
+
+# Use the same function as previous part to extract the nuclei masks from pre-trained cellpose model.
+def cellpose_segmentation(prediction:ArrayLike,target:ArrayLike)->Tuple[torch.ShortTensor]:
+ # NOTE these are hardcoded for this notebook and A549 dataset
+ device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
+ cp_nuc_kwargs = {
+ "diameter": 65,
+ "cellprob_threshold": 0.0,
+ }
+ cellpose_model = models.CellposeModel(
+ gpu=True, model_type='nuclei', device=torch.device(device)
+ )
+ pred_label, _, _ = cellpose_model.eval(prediction, **cp_nuc_kwargs)
+ target_label, _, _ = cellpose_model.eval(target, **cp_nuc_kwargs)
+
+ pred_label = pred_label.astype(np.int32)
+ target_label = target_label.astype(np.int32)
+ pred_label = torch.ShortTensor(pred_label)
+ target_label = torch.ShortTensor(target_label)
+
+ return (pred_label,target_label)
+
+# Define dataframe to store the segmentation metrics.
+test_segmentation_metrics= pd.DataFrame(
+ columns=["model", "fov","masks_per_fov","accuracy","dice","jaccard","mAP","mAP_50","mAP_75","mAR_100"]
+)
+# Define tuple to store the segmentation results. Each value in the tuple is a dictionary containing the model name, fov, predicted label, predicted stain, target label, and target stain.
+segmentation_results = ()
+
+for i, (target_stain, predicted_stain) in tqdm(enumerate(zip(target_stains, virtual_stains)),desc='Computing Metrics',total=len(target_stains)):
+ fov = str(virtual_stain_paths[i]).split("/")[-1].split(".")[0]
+ minmax_norm_target = min_max_scale(target_stain)
+ minmax_norm_predicted = min_max_scale(predicted_stain)
+ # Compute the segmentation masks.
+ pred_label, target_label = cellpose_segmentation(minmax_norm_predicted, minmax_norm_target)
+ # Binary labels
+ pred_label_binary = pred_label > 0
+ target_label_binary = target_label > 0
+
+ # Use Coco metrics to get mean average precision
+ coco_metrics = mean_average_precision(pred_label, target_label)
+ # Find unique number of labels
+ num_masks_fov = len(np.unique(pred_label))
+ # Find unique number of labels
+ num_masks_fov = len(np.unique(pred_label))
+ # Compute the segmentation metrics.
+ test_segmentation_metrics.loc[len(test_segmentation_metrics)] = {
+ "model": "pix2pixHD",
+ "fov":fov,
+ "masks_per_fov": num_masks_fov,
+ "accuracy": accuracy(pred_label_binary, target_label_binary, task="binary").item(),
+ "dice": dice(pred_label_binary, target_label_binary).item(),
+ "jaccard": jaccard_index(pred_label_binary, target_label_binary, task="binary").item(),
+ "mAP":coco_metrics["map"].item(),
+ "mAP_50":coco_metrics["map_50"].item(),
+ "mAP_75":coco_metrics["map_75"].item(),
+ "mAR_100":coco_metrics["mar_100"].item()
+ }
+ # Store the segmentation results.
+ segmentation_result = {
+ "model": "pix2pixHD",
+ "fov":fov,
+ "phase_image": phase_images[i],
+ "pred_label": pred_label,
+ "pred_stain": predicted_stain,
+ "target_label": target_label,
+ "target_stain": target_stain,
+ }
+ segmentation_results += (segmentation_result,)
+
+test_segmentation_metrics.head()
+# %%
+# Define function to visualize the segmentation results.
+def visualise_results_and_masks(segmentation_results: Tuple[dict], segmentation_metrics: pd.DataFrame, rows: int = 5, crop_size: int = None, crop_type: str = 'center'):
+
+ # Sample a subset of the segmentation results.
+ sample_indices = np.random.choice(len(phase_images),rows)
+ print(sample_indices)
+ segmentation_metrics = segmentation_metrics.iloc[sample_indices,:]
+ segmentation_results = [segmentation_results[i] for i in sample_indices]
+ # Define the figure and axes.
+ fig, axes = plt.subplots(rows, 5, figsize=(rows*3, 15))
+
+ # Visualize the segmentation results.
+ for i in range(len((segmentation_results))):
+ segmentation_metric = segmentation_metrics.iloc[i]
+ result = segmentation_results[i]
+ phase_image = result["phase_image"]
+ target_stain = result["target_stain"]
+ target_label = result["target_label"]
+ pred_stain = result["pred_stain"]
+ pred_label = result["pred_label"]
+ # Crop the images if required. Zoom into instances
+ if crop_size is not None:
+ phase_image = crop(phase_image, crop_size, crop_type)
+ target_stain = crop(target_stain, crop_size, crop_type)
+ target_label = crop(target_label, crop_size, crop_type)
+ pred_stain = crop(pred_stain, crop_size, crop_type)
+ pred_label = crop(pred_label, crop_size, crop_type)
+
+ axes[i, 0].imshow(phase_image, cmap="gray")
+ axes[i, 0].set_title("Phase")
+ axes[i, 1].imshow(
+ target_stain,
+ cmap="gray",
+ vmin=np.percentile(target_stain, 1),
+ vmax=np.percentile(target_stain, 99),
+ )
+ axes[i, 1].set_title("Target Fluorescence")
+ axes[i, 2].imshow(pred_stain, cmap="gray")
+ axes[i, 2].set_title("Virtual Stain")
+ axes[i, 3].imshow(target_label, cmap="inferno")
+ axes[i, 3].set_title("Target Fluorescence Mask")
+ axes[i, 4].imshow(pred_label, cmap="inferno")
+ # Add Metric values to the title
+ axes[i, 4].set_title(f"Virtual Stain Mask\nAcc:{segmentation_metric['accuracy']:.2f} Dice:{segmentation_metric['dice']:.2f}\nJaccard:{segmentation_metric['jaccard']:.2f} MAP:{segmentation_metric['mAP']:.2f}")
+ # Turn off the axes.
+ for ax in axes.flatten():
+ ax.axis("off")
+
+ plt.tight_layout()
+ plt.show()
+
+visualise_results_and_masks(segmentation_results,test_segmentation_metrics, crop_size=256, crop_type='center')
+
+# %% [markdown]
+# %% [markdown]
+"""
+## Inference Instance-level Results
+Please note down your thoughts about the following questions...
+
+- What do these metrics tells us about the performance of the model?
+- How does the performance compare to when looking at pixel-level metrics?
+
+"""
+# %% [markdown]
+"""
+
+
+## Checkpoint 3
+
+Congratulations! You have generated predictions from a pre-trained model and evaluated the performance of the model on unseen data. You have computed pixel-level metrics and instance-level metrics to evaluate the performance of the model. You may have also began training your own Pix2PixHD GAN models with alternative hyperparameters.
+
+
+"""
+
+# %% [markdown]
+"""
+# Part 4. Visualise Regression vs Generative Modelling Approaches
+--------------------------------------------------
+"""
+# %% tags=["task"]
+# Load Viscy Virtual Stains
+viscy_results_path = "/ADD/PATH/TO/RESULTS/HERE"
+viscy_stain_paths = sorted([i for i in Path(viscy_results_path).glob("**/*.tiff")])
+assert len(viscy_stain_paths) == len(virtual_stain_paths), "Number of images do not match."
+visy_stains = np.zeros((len(viscy_stain_paths), 512, 512))
+for index, v_path in enumerate(viscy_stain_paths):
+ viscy_stain = imread(v_path)
+ visy_stains[index] = viscy_stain
+
+##########################
+######## TODO ########
+##########################
+
+
+def visualise_both_methods():
+ # Your code here
+ pass
+
+
+# %% tags=["solution"]
+
+##########################
+######## Solution ########
+##########################
+
+def visualise_both_methods(
+ phase_images: np.array, target_stains: np.array, pix2pixHD_results: np.array, viscy_results: np.array,crop_size=None,crop_type='center'
+):
+ fig, axes = plt.subplots(5, 4, figsize=(15, 15))
+ sample_indices = np.random.choice(len(phase_images), 5)
+ if crop_size is not None:
+ phase_image = crop(phase_image, crop_size, crop_type)
+ target_stain = crop(target_stain, crop_size, crop_type)
+ target_label = crop(target_label, crop_size, crop_type)
+ pred_stain = crop(pred_stain, crop_size, crop_type)
+ pred_label = crop(pred_label, crop_size, crop_type)
+
+ for i, idx in enumerate(sample_indices):
+ axes[i, 0].imshow(phase_images[idx], cmap="gray")
+ axes[i, 0].set_title("Phase")
+ axes[i, 0].axis("off")
+ axes[i, 1].imshow(
+ target_stains[idx],
+ cmap="gray",
+ vmin=np.percentile(target_stains[idx], 1),
+ vmax=np.percentile(target_stains[idx], 99),
+ )
+ axes[i, 1].set_title("Nuclei")
+ axes[i, 1].axis("off")
+
+ axes[i, 2].imshow(
+ viscy_results[idx],
+ cmap="gray",
+ vmin=np.percentile(target_stains[idx], 1),
+ vmax=np.percentile(target_stains[idx], 99),
+ )
+ axes[i, 2].set_title("Regression\nVirtual Stain")
+ axes[i, 2].axis("off")
+
+ axes[i, 3].imshow(
+ pix2pixHD_results[idx],
+ cmap="gray",
+ vmin=np.percentile(target_stains[idx], 1),
+ vmax=np.percentile(target_stains[idx], 99),
+ )
+ axes[i, 3].set_title("Pix2PixHD GAN\nVirtual Stain")
+ axes[i, 3].axis("off")
+ plt.tight_layout()
+ plt.show()
+# %% [markdown]
+"""
+
+
+## Checkpoint 4
+
+Congratulations! You should now have a better understanding of the difference in performance for image translation when approaching the problem using a regression vs. generative modelling approaches!
+
+
+"""
+# %% [markdown]
+"""
+# Part 5: BONUS: Sample different virtual staining solutions from the GAN using MC-Dropout and explore the uncertainty in the virtual stain predictions.
+--------------------------------------------------
+Steps:
+- Load the pre-trained model.
+- Generate multiple predictions for the same input image.
+- Compute the pixel-wise variance across the predictions.
+- Visualise the pixel-wise variance to explore the uncertainty in the virtual stain predictions.
+
+"""
+# %%
+# Use the same model and dataloaders as before.
+# Load the test data.
+test_data_loader = CreateDataLoader(opt)
+test_dataset = test_data_loader.load_data()
+visualizer = Visualizer(opt)
+
+# Load pre-trained model
+opt.variational_inf_runs = 100 # Number of samples per phase input
+opt.variation_inf_path = f"./GAN_code/GANs_MI2I/pre_trained/{opt.name}/samples/" # Path to store the samples.
+opt.results_dir = f"{top_dir}/GAN_code/GANs_MI2I/pre_trained/dlmbl_vsnuclei/sampling_results"
+opt.dropout_variation_inf = True # Use dropout during inference.
+model = create_model(opt)
+# Generate & save predictions in the variation_inf_path directory.
+sampling(test_dataset, opt, model)
+
+# %%
+# Visualise Samples
+samples = sorted([i for i in Path(f"{opt.results_dir}").glob("**/*.tif*")])
+assert len(samples) == 5
+# Create arrays to store the images.
+sample_images = np.zeros((5,100, 512, 512)) # (samples, images, height, width)
+# Load the images and store them in the arrays.
+for index, sample_path in tqdm(enumerate(samples)):
+ sample_image = imread(sample_path)
+ # Append the images to the arrays.
+ sample_images[index] = sample_image
+
+# %%
+# Create a matplotlib plot with animation through images.
+def animate_images(images):
+ # Expecting images to have shape (frames, height, width)
+ fig, ax = plt.subplots()
+ ax.axis('off')
+
+ # Make sure images are in (frames, height, width) order
+ images = images.transpose(0, 2, 1) if images.shape[1] == images.shape[2] else images
+
+ imgs = []
+ for i in range(min(100, len(images))): # Ensure you don't exceed the number of frames
+ im = ax.imshow(images[i], animated=True)
+ imgs.append([im])
+
+ ani = animation.ArtistAnimation(fig, imgs, interval=100, blit=False, repeat_delay=1000)
+
+ # Display the animation
+ # plt.close(fig)
+ display(HTML(ani.to_jshtml()))
+
+# Example call with sample_images[0]
+animate_images(sample_images[0])
+
+# %% [markdown]
+"""
+
+
+## Checkpoint 5
+
+Congratulations! This is the end of the conditional generative modelling approach to image translation notebook. You have trained and examined the loss components of Pix2PixHD GAN. You have compared the results of a regression-based approach vs. generative modelling approach and explored the variability in virtual staining solutions. I hope you have enjoyed learning experience!
+
+"""
diff --git a/setup.sh b/setup.sh
deleted file mode 100644
index 9472b16..0000000
--- a/setup.sh
+++ /dev/null
@@ -1,32 +0,0 @@
-#!/usr/bin/env -S bash -i
-
-START_DIR=$(pwd)
-
-# Create mamba environment
-mamba create -y --name 04_image_translation python=3.10
-
-# Install ipykernel in the environment.
-mamba install -y ipykernel nbformat nbconvert black jupytext ipywidgets --name 04_image_translation
-# Specifying the environment explicitly.
-# mamba activate sometimes doesn't work from within shell scripts.
-
-# install viscy and its dependencies`s in the environment using pip.
-mkdir -p ~/code/
-cd ~/code/
-git clone https://github.com/mehta-lab/viscy.git
-cd viscy
-git checkout 7c5e4c1d68e70163cf514d22c475da8ea7dc3a88 # Exercise is tested with this commit of viscy
-# Find path to the environment - mamba activate doesn't work from within shell scripts.
-ENV_PATH=$(conda info --envs | grep 04_image_translation | awk '{print $NF}')
-$ENV_PATH/bin/pip install ."[metrics]"
-
-# Create data directory
-mkdir -p ~/data/04_image_translation
-cd ~/data/04_image_translation
-wget https://dl-at-mbl-2023-data.s3.us-east-2.amazonaws.com/DLMBL2023_image_translation_data_pyramid.tar.gz
-wget https://dl-at-mbl-2023-data.s3.us-east-2.amazonaws.com/DLMBL2023_image_translation_test.tar.gz
-tar -xzf DLMBL2023_image_translation_data_pyramid.tar.gz
-tar -xzf DLMBL2023_image_translation_test.tar.gz
-
-# Change back to the starting directory
-cd $START_DIR
diff --git a/solution.ipynb b/solution.ipynb
deleted file mode 100644
index 5775c64..0000000
--- a/solution.ipynb
+++ /dev/null
@@ -1,1314 +0,0 @@
-{
- "cells": [
- {
- "cell_type": "markdown",
- "id": "15d23751",
- "metadata": {
- "cell_marker": "\"\"\""
- },
- "source": [
- "# Image translation\n",
- "---\n",
- "\n",
- "Written by Ziwen Liu and Shalin Mehta, CZ Biohub San Francisco.\n",
- "\n",
- "In this exercise, we will solve an image translation task to predict fluorescence images of nuclei and membrane markers from quantitative phase images of cells. In other words, we will _virtually stain_ the nuclei and membrane visible in the phase image. \n",
- "\n",
- "Here, the source domain is label-free microscopy (material density) and the target domain is fluorescence microscopy (fluorophore density). The goal is to learn a mapping from the source domain to the target domain. We will use a deep convolutional neural network (CNN), specifically, a U-Net model with residual connections to learn the mapping. The preprocessing, training, prediction, evaluation, and deployment steps are unified in a computer vision pipeline for single-cell analysis that we call [VisCy](https://github.com/mehta-lab/VisCy).\n",
- "\n",
- "VisCy evolved from our previous work on virtual staining of cellular components from their density and anisotropy.\n",
- "\n",
- "\n",
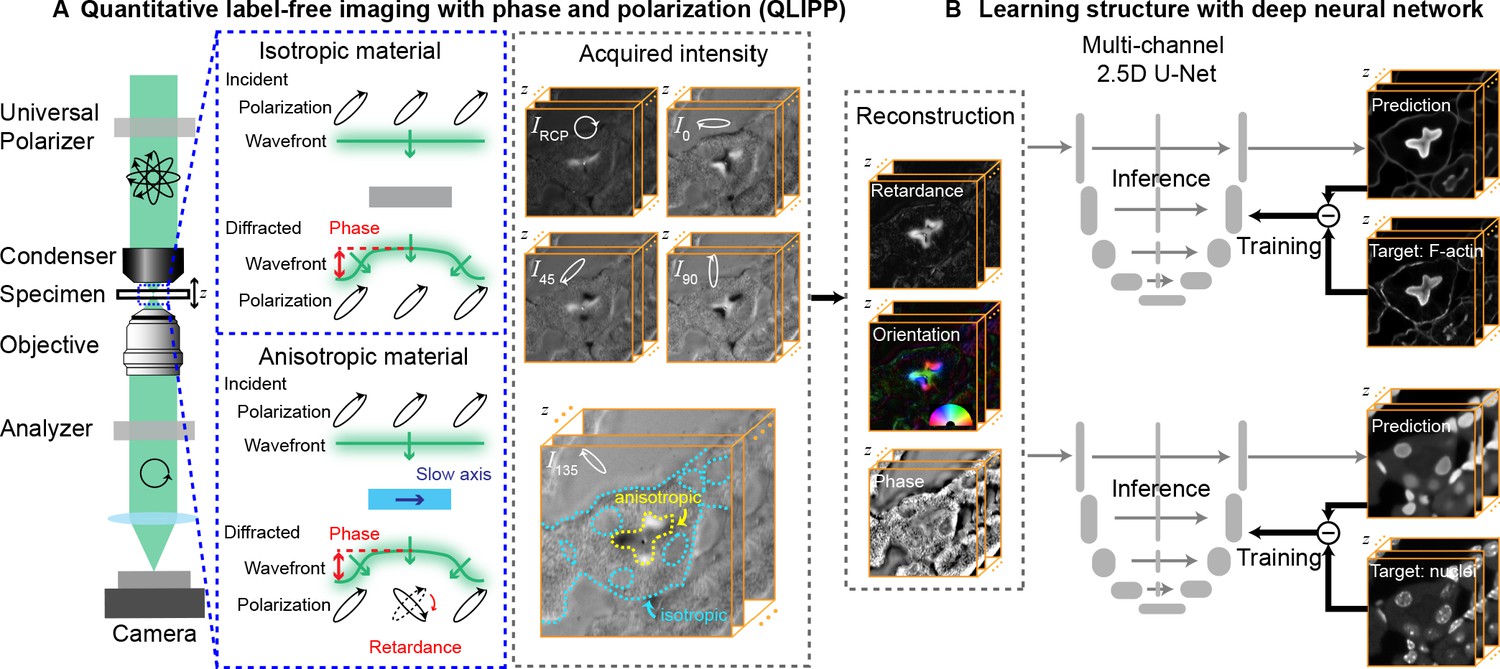
- "[Guo et al. (2020) Revealing architectural order with quantitative label-free imaging and deep learning\n",
- ". eLife](https://elifesciences.org/articles/55502).\n",
- "\n",
- "VisCy exploits recent advances in the data and metadata formats ([OME-zarr](https://www.nature.com/articles/s41592-021-01326-w)) and DL frameworks, [PyTorch Lightning](https://lightning.ai/) and [MONAI](https://monai.io/). "
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b5957320",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "Today, we will train a 2D image translation model using a 2D U-Net with residual connections. We will use a dataset of 301 fields of view (FOVs) of Human Embryonic Kidney (HEK) cells, each FOV has 3 channels (phase, membrane, and nuclei). The cells were labeled with CRISPR editing. Intrestingly, not all cells during this experiment were labeled due to the stochastic nature of CRISPR editing. In such situations, virtual staining rescues missing labels.\n",
- ""
- ]
- },
- {
- "cell_type": "markdown",
- "id": "6c62db93",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "
\n",
- "The exercise is organized in 3 parts.\n",
- "\n",
- "* **Part 1** - Explore the data using tensorboard. Launch the training before lunch.\n",
- "* Lunch break - The model will continue training during lunch.\n",
- "* **Part 2** - Evaluate the training with tensorboard. Train another model.\n",
- "* **Part 3** - Tune the models to improve performance.\n",
- "
"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "30739bb5",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "📖 As you work through parts 2 and 3, please share the layouts of your models (output of torchview) and their performance with everyone via [this google doc](https://docs.google.com/document/d/1hZWSVRvt9KJEdYu7ib-vFBqAVQRYL8cWaP_vFznu7D8/edit#heading=h.n5u485pmzv2z) 📖.\n",
- "\n",
- "\n",
- "Our guesstimate is that each of the three parts will take ~1.5 hours. A reasonable 2D UNet can be trained in ~20 min on a typical AWS node. \n",
- "We will discuss your observations on google doc after checkpoints 2 and 3.\n",
- "\n",
- "The focus of the exercise is on understanding information content of the data, how to train and evaluate 2D image translation model, and explore some hyperparameters of the model. If you complete this exercise and have time to spare, try the bonus exercise on 3D image translation."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "658e3b31",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "
\n",
- "Set your python kernel to 04_image_translation\n",
- "
"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "7b26db60",
- "metadata": {
- "cell_marker": "\"\"\"",
- "incorrectly_encoded_metadata": "id='1_phase2fluor'>",
- "title": "
\n",
- "If you launched jupyter lab from ssh terminal, add --host <your-server-name> to the tensorboard command below. <your-server-name> is the address of your compute node that ends in amazonaws.com.\n",
- "\n",
- "You can also launch tensorboard in an independent tab (instead of in the notebook) by changing the `%` to `!`\n",
- "\n",
- "\n",
- "### Task 1.1\n",
- " \n",
- "Look at a couple different fields of view by changing the value in the cell above. See if you notice any missing or inconsistent staining.\n",
- "
"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "7f6f3609",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 1
- },
- "source": [
- "## Explore the effects of augmentation on batch.\n",
- "\n",
- "VisCy builds on top of PyTorch Lightning. PyTorch Lightning is a thin wrapper around PyTorch that allows rapid experimentation. It provides a [DataModule](https://lightning.ai/docs/pytorch/stable/data/datamodule.html) to handle loading and processing of data during training. VisCy provides a child class, `HCSDataModule` to make it intuitve to access data stored in the HCS layout.\n",
- " \n",
- "The dataloader in `HCSDataModule` returns a batch of samples. A `batch` is a list of dictionaries. The length of the list is equal to the batch size. Each dictionary consists of following key-value pairs.\n",
- "- `source`: the input image, a tensor of size 1*1*Y*X\n",
- "- `target`: the target image, a tensor of size 2*1*Y*X\n",
- "- `index` : the tuple of (location of field in HCS layout, time, and z-slice) of the sample."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "1684d72e",
- "metadata": {},
- "source": [
- "\n",
- "\n",
- "### Task 1.2\n",
- "\n",
- "Setup the data loader and log several batches to tensorboard.\n",
- "\n",
- "Based on the tensorboard images, what are the two channels in the target image?\n",
- "\n",
- "Note: If tensorboard is not showing images, try refreshing and using the \"Images\" tab.\n",
- "
"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "67211280",
- "metadata": {},
- "outputs": [],
- "source": [
- "# Define a function to write a batch to tensorboard log.\n",
- "\n",
- "def log_batch_tensorboard(batch, batchno, writer, card_name):\n",
- " \"\"\"\n",
- " Logs a batch of images to TensorBoard.\n",
- "\n",
- " Args:\n",
- " batch (dict): A dictionary containing the batch of images to be logged.\n",
- " writer (SummaryWriter): A TensorBoard SummaryWriter object.\n",
- " card_name (str): The name of the card to be displayed in TensorBoard.\n",
- "\n",
- " Returns:\n",
- " None\n",
- " \"\"\"\n",
- " batch_phase = batch[\"source\"][:, :, 0, :, :] # batch_size x z_size x Y x X tensor.\n",
- " batch_membrane = batch[\"target\"][:, 1, 0, :, :].unsqueeze(\n",
- " 1\n",
- " ) # batch_size x 1 x Y x X tensor.\n",
- " batch_nuclei = batch[\"target\"][:, 0, 0, :, :].unsqueeze(\n",
- " 1\n",
- " ) # batch_size x 1 x Y x X tensor.\n",
- "\n",
- " p1, p99 = np.percentile(batch_membrane, (0.1, 99.9))\n",
- " batch_membrane = np.clip((batch_membrane - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " p1, p99 = np.percentile(batch_nuclei, (0.1, 99.9))\n",
- " batch_nuclei = np.clip((batch_nuclei - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " p1, p99 = np.percentile(batch_phase, (0.1, 99.9))\n",
- " batch_phase = np.clip((batch_phase - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " [N, C, H, W] = batch_phase.shape\n",
- " interleaved_images = torch.zeros((3 * N, C, H, W), dtype=batch_phase.dtype)\n",
- " interleaved_images[0::3, :] = batch_phase\n",
- " interleaved_images[1::3, :] = batch_nuclei\n",
- " interleaved_images[2::3, :] = batch_membrane\n",
- "\n",
- " grid = torchvision.utils.make_grid(interleaved_images, nrow=3)\n",
- "\n",
- " # add the grid to tensorboard\n",
- " writer.add_image(card_name, grid, batchno)"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "73577e5a",
- "metadata": {},
- "outputs": [],
- "source": [
- "# Define a function to visualize a batch on jupyter, in case tensorboard is finicky \n",
- "\n",
- "def log_batch_jupyter(batch):\n",
- " \"\"\"\n",
- " Logs a batch of images on jupyter using ipywidget.\n",
- "\n",
- " Args:\n",
- " batch (dict): A dictionary containing the batch of images to be logged.\n",
- "\n",
- " Returns:\n",
- " None\n",
- " \"\"\"\n",
- " batch_phase = batch[\"source\"][:, :, 0, :, :] # batch_size x z_size x Y x X tensor.\n",
- " batch_size = batch_phase.shape[0]\n",
- " batch_membrane = batch[\"target\"][:, 1, 0, :, :].unsqueeze(\n",
- " 1\n",
- " ) # batch_size x 1 x Y x X tensor.\n",
- " batch_nuclei = batch[\"target\"][:, 0, 0, :, :].unsqueeze(\n",
- " 1\n",
- " ) # batch_size x 1 x Y x X tensor.\n",
- "\n",
- " p1, p99 = np.percentile(batch_membrane, (0.1, 99.9))\n",
- " batch_membrane = np.clip((batch_membrane - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " p1, p99 = np.percentile(batch_nuclei, (0.1, 99.9))\n",
- " batch_nuclei = np.clip((batch_nuclei - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " p1, p99 = np.percentile(batch_phase, (0.1, 99.9))\n",
- " batch_phase = np.clip((batch_phase - p1) / (p99 - p1), 0, 1)\n",
- "\n",
- " plt.figure()\n",
- " fig, axes = plt.subplots(batch_size, n_channels, figsize=(10, 10))\n",
- " [N, C, H, W] = batch_phase.shape\n",
- " for sample_id in range(batch_size):\n",
- " axes[sample_id, 0].imshow(batch_phase[sample_id,0])\n",
- " axes[sample_id, 1].imshow(batch_nuclei[sample_id,0])\n",
- " axes[sample_id, 2].imshow(batch_membrane[sample_id,0])\n",
- "\n",
- " for i in range(n_channels):\n",
- " axes[sample_id, i].axis(\"off\")\n",
- " axes[sample_id, i].set_title(dataset.channel_names[i])\n",
- " plt.tight_layout()\n",
- " plt.show()"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "398f4546",
- "metadata": {
- "lines_to_next_cell": 2
- },
- "outputs": [],
- "source": [
- "\n",
- "# Initialize the data module.\n",
- "\n",
- "BATCH_SIZE = 4\n",
- "# 42 is a perfectly reasonable batch size. After all, it is the answer to the ultimate question of life, the universe and everything.\n",
- "# More seriously, batch size does not have to be a power of 2.\n",
- "# See: https://sebastianraschka.com/blog/2022/batch-size-2.html\n",
- "\n",
- "data_module = HCSDataModule(\n",
- " data_path,\n",
- " source_channel=\"Phase\",\n",
- " target_channel=[\"Nuclei\", \"Membrane\"],\n",
- " z_window_size=1,\n",
- " split_ratio=0.8,\n",
- " batch_size=BATCH_SIZE,\n",
- " num_workers=8,\n",
- " architecture=\"2D\",\n",
- " yx_patch_size=(512, 512), # larger patch size makes it easy to see augmentations.\n",
- " augment=False, # Turn off augmentation for now.\n",
- ")\n",
- "data_module.setup(\"fit\")\n",
- "\n",
- "print(\n",
- " f\"FOVs in training set: {len(data_module.train_dataset)}, FOVs in validation set:{len(data_module.val_dataset)}\"\n",
- ")\n",
- "train_dataloader = data_module.train_dataloader()\n",
- "\n",
- "# Instantiate the tensorboard SummaryWriter, logs the first batch and then iterates through all the batches and logs them to tensorboard.\n",
- "\n",
- "writer = SummaryWriter(log_dir=f\"{log_dir}/view_batch\")\n",
- "# Draw a batch and write to tensorboard.\n",
- "batch = next(iter(train_dataloader))\n",
- "log_batch_tensorboard(batch, 0, writer, \"augmentation/none\")\n",
- "writer.close()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "a4c45450",
- "metadata": {},
- "source": [
- "Visualize directly on Jupyter ☄️, if your tensorboard is causing issues."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "73466d3f",
- "metadata": {},
- "outputs": [],
- "source": [
- "%matplotlib inline\n",
- "log_batch_jupyter(batch)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "19def8d6",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "## View augmentations using tensorboard."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "97bdcbd8",
- "metadata": {},
- "outputs": [],
- "source": [
- "# Here we turn on data augmentation and rerun setup\n",
- "data_module.augment = True\n",
- "data_module.setup(\"fit\")\n",
- "\n",
- "# get the new data loader with augmentation turned on\n",
- "augmented_train_dataloader = data_module.train_dataloader()\n",
- "\n",
- "# Draw batches and write to tensorboard\n",
- "writer = SummaryWriter(log_dir=f\"{log_dir}/view_batch\")\n",
- "augmented_batch = next(iter(augmented_train_dataloader))\n",
- "log_batch_tensorboard(augmented_batch, 0, writer, \"augmentation/some\")\n",
- "writer.close()"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "247bf9c7",
- "metadata": {},
- "source": [
- "Visualize directly on Jupyter ☄️"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "de281de7",
- "metadata": {},
- "outputs": [],
- "source": [
- "log_batch_jupyter(augmented_batch)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "47cc91e4",
- "metadata": {},
- "source": [
- "\n",
- "\n",
- "### Task 1.3\n",
- "Can you tell what augmentation were applied from looking at the augmented images in Tensorboard?\n",
- "\n",
- "Check your answer using the source code [here](https://github.com/mehta-lab/VisCy/blob/b89f778b34735553cf155904eef134c756708ff2/viscy/light/data.py#L529).\n",
- "
"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "5d05336e",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "## Train a 2D U-Net model to predict nuclei and membrane from phase.\n",
- "\n",
- "### Construct a 2D U-Net\n",
- "See ``viscy.unet.networks.Unet2D.Unet2d`` ([source code](https://github.com/mehta-lab/VisCy/blob/7c5e4c1d68e70163cf514d22c475da8ea7dc3a88/viscy/unet/networks/Unet2D.py#L7)) for configuration details."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "964d5aae",
- "metadata": {
- "lines_to_next_cell": 2
- },
- "outputs": [],
- "source": [
- "# Create a 2D UNet.\n",
- "GPU_ID = 0\n",
- "BATCH_SIZE = 10\n",
- "YX_PATCH_SIZE = (512, 512)\n",
- "\n",
- "\n",
- "# Dictionary that specifies key parameters of the model.\n",
- "phase2fluor_config = {\n",
- " \"architecture\": \"2D\",\n",
- " \"num_filters\": [24, 48, 96, 192, 384],\n",
- " \"in_channels\": 1,\n",
- " \"out_channels\": 2,\n",
- " \"residual\": True,\n",
- " \"dropout\": 0.1, # dropout randomly turns off weights to avoid overfitting of the model to data.\n",
- " \"task\": \"reg\", # reg = regression task.\n",
- "}\n",
- "\n",
- "phase2fluor_model = VSUNet(\n",
- " model_config=phase2fluor_config.copy(),\n",
- " batch_size=BATCH_SIZE,\n",
- " loss_function=torch.nn.functional.l1_loss,\n",
- " schedule=\"WarmupCosine\",\n",
- " log_num_samples=5, # Number of samples from each batch to log to tensorboard.\n",
- " example_input_yx_shape=YX_PATCH_SIZE,\n",
- ")"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "eabb4902",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 0
- },
- "source": [
- "### Instantiate data module and trainer, test that we are setup to launch training."
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "9728a1c0",
- "metadata": {
- "lines_to_next_cell": 2
- },
- "outputs": [],
- "source": [
- "# Setup the data module.\n",
- "phase2fluor_data = HCSDataModule(\n",
- " data_path,\n",
- " source_channel=\"Phase\",\n",
- " target_channel=[\"Nuclei\", \"Membrane\"],\n",
- " z_window_size=1,\n",
- " split_ratio=0.8,\n",
- " batch_size=BATCH_SIZE,\n",
- " num_workers=8,\n",
- " architecture=\"2D\",\n",
- " yx_patch_size=YX_PATCH_SIZE,\n",
- " augment=True,\n",
- ")\n",
- "phase2fluor_data.setup(\"fit\")\n",
- "# fast_dev_run runs a single batch of data through the model to check for errors.\n",
- "trainer = VSTrainer(accelerator=\"gpu\", devices=[GPU_ID], fast_dev_run=True)\n",
- "\n",
- "# trainer class takes the model and the data module as inputs.\n",
- "trainer.fit(phase2fluor_model, datamodule=phase2fluor_data)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "b837c6b2",
- "metadata": {},
- "source": [
- "## View model graph.\n",
- "\n",
- "PyTorch uses dynamic graphs under the hood. The graphs are constructed on the fly. This is in contrast to TensorFlow, where the graph is constructed before the training loop and remains static. In other words, the graph of the network can change with every forward pass. Therefore, we need to supply an input tensor to construct the graph. The input tensor can be a random tensor of the correct shape and type. We can also supply a real image from the dataset. The latter is more useful for debugging."
- ]
- },
- {
- "cell_type": "markdown",
- "id": "31665d0f",
- "metadata": {},
- "source": [
- "\n",
- "\n",
- "### Task 1.4\n",
- "Run the next cell to generate a graph representation of the model architecture. Can you recognize the UNet structure and skip connections in this graph visualization?\n",
- "
"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "ac20e2d3",
- "metadata": {},
- "outputs": [],
- "source": [
- "# visualize graph of phase2fluor model as image.\n",
- "model_graph_phase2fluor = torchview.draw_graph(\n",
- " phase2fluor_model,\n",
- " phase2fluor_data.train_dataset[0][\"source\"],\n",
- " depth=2, # adjust depth to zoom in.\n",
- " device=\"cpu\",\n",
- ")\n",
- "# Print the image of the model.\n",
- "model_graph_phase2fluor.visual_graph"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "496019cb",
- "metadata": {
- "cell_marker": "\"\"\"",
- "lines_to_next_cell": 2
- },
- "source": [
- "\n",
- "\n",
- "### Task 1.5\n",
- "Start training by running the following cell. Check the new logs on the tensorboard.\n",
- "
"
- ]
- },
- {
- "cell_type": "code",
- "execution_count": null,
- "id": "26303693",
- "metadata": {},
- "outputs": [],
- "source": [
- "\n",
- "GPU_ID = 0\n",
- "n_samples = len(phase2fluor_data.train_dataset)\n",
- "steps_per_epoch = n_samples // BATCH_SIZE # steps per epoch.\n",
- "n_epochs = 50 # Set this to 50 or the number of epochs you want to train for.\n",
- "\n",
- "trainer = VSTrainer(\n",
- " accelerator=\"gpu\",\n",
- " devices=[GPU_ID],\n",
- " max_epochs=n_epochs,\n",
- " log_every_n_steps=steps_per_epoch // 2,\n",
- " # log losses and image samples 2 times per epoch.\n",
- " logger=TensorBoardLogger(\n",
- " save_dir=log_dir,\n",
- " # lightning trainer transparently saves logs and model checkpoints in this directory.\n",
- " name=\"phase2fluor\",\n",
- " log_graph=True,\n",
- " ),\n",
- " ) \n",
- "# Launch training and check that loss and images are being logged on tensorboard.\n",
- "trainer.fit(phase2fluor_model, datamodule=phase2fluor_data)"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "4260177d",
- "metadata": {
- "cell_marker": "\"\"\""
- },
- "source": [
- "\n",
- "\n",
- "## Checkpoint 1\n",
- "\n",
- "Now the training has started,\n",
- "we can come back after a while and evaluate the performance!\n",
- "
"
- ]
- },
- {
- "cell_type": "markdown",
- "id": "eb238ae2",
- "metadata": {
- "cell_marker": "\"\"\"",
- "incorrectly_encoded_metadata": "id='1_fluor2phase'>",
- "title": "