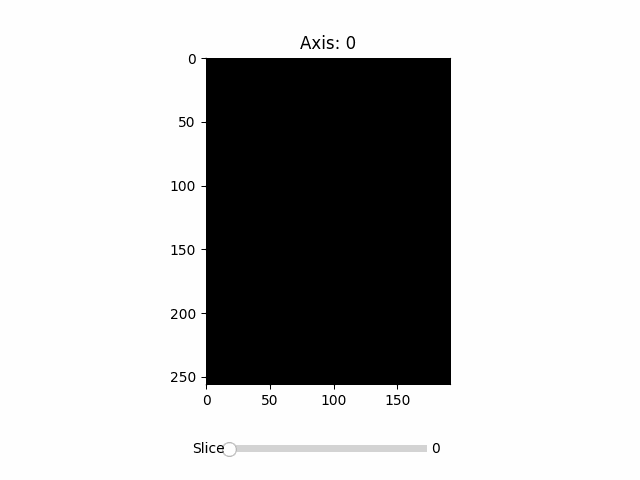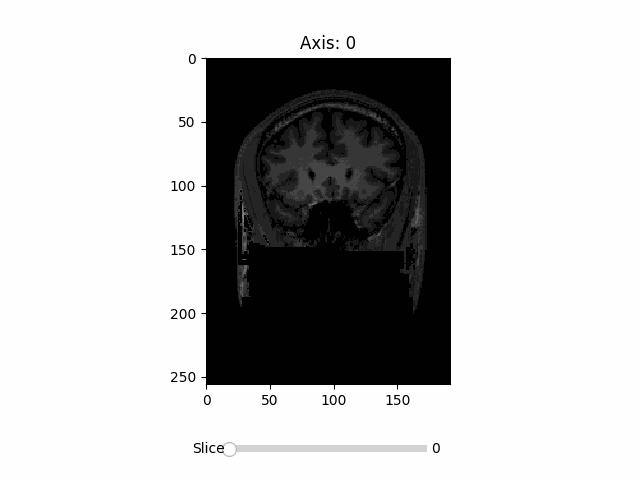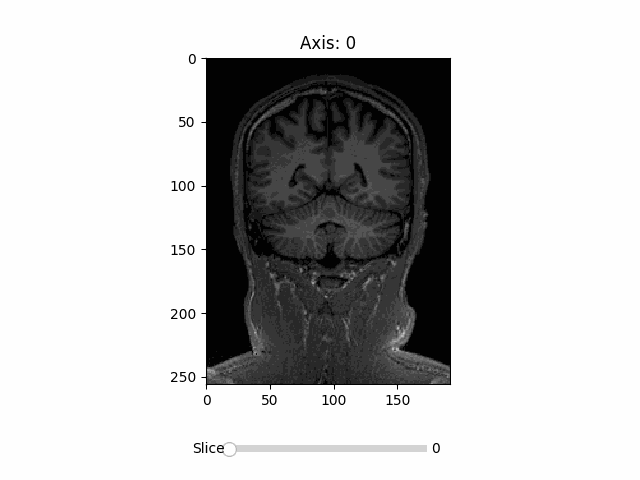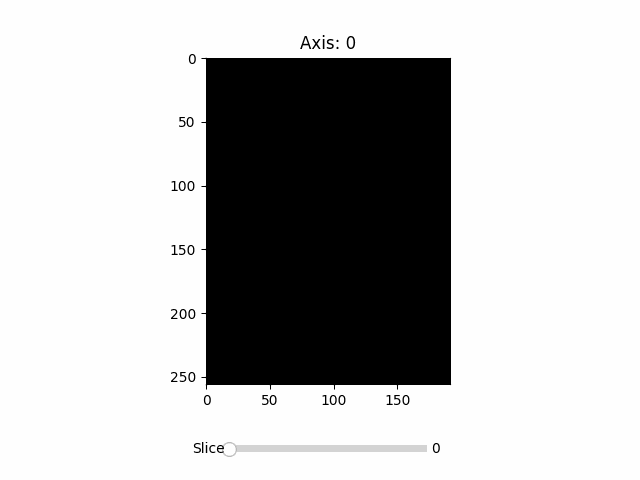
🌴 RadVis (Radiology Visualization) is a simple visualization package for medical imaging 🌴
pip install radvis
Loading an image and displaying it with a slider
import radvis as rv
# Creates a 'RadImage' object containing the image data
image = rv.load_image('path/to/image.nii.gz') # Can also load from DICOM or Numpy files
slicer = rv.RadSlicer(image, axis=0)
slicer.display()Create a RadImage from a numpy array
import radvis as rv
import numpy as np
# Creating a numpy array
image_data = np.random.rand(100, 100, 100)
# Creating a 'RadImage' object from the numpy array
image = rv.from_numpy(image_data)
slicer = rv.RadSlicer(image, axis=0)
slicer.display()Adding masks to the image
import radvis as rv
import numpy as np
image = rv.load_image('path/to/image.nii.gz')
slicer = rv.RadSlicer(image, axis=0)
# 0 values wont be displayed
red_mask = np.zeros_like(image.image_data)
red_mask[5:100, 5:100, 5:100] = 1
blue_mask = np.zeros_like(image.image_data)
blue_mask[70:150, 70:150, 70:150] = 1
slicer.add_mask(red_mask, color="red")
slicer.add_mask(blue_mask, color="blue")
slicer.display()Mask can be another RadImage object so you can load up your masks from a DICOM or NIFTI
import radvis as rv
import numpy as np
AXIS = 1
IMAGE_PATH = "path/to/image.nii.gz"
IMAGE_MASK_PATH = "path/to/mask.nii.gz"
image = rv.load_image(IMAGE_PATH)
mask = rv.load_image(IMAGE_MASK_PATH)
slicer = rv.RadSlicer(image, AXIS, width=3)
slicer.add_mask(mask, color="red", alpha=0.3)
slicer.display()
slicer.save_animation(f"images/axis_{AXIS}_brain_seg.gif", fps=30)
slicer.save_frame(f"images/axis_{AXIS}_brain_seg.png", index=180, dpi=300)You can also display multiple slicers at once 🤯
import radvis as rv
import numpy as np
img1 = rv.from_numpy(np.random.rand(50, 10, 10))
img2 = rv.from_numpy(np.random.rand(10, 50, 10))
img3 = rv.from_numpy(np.random.rand(10, 10, 50))
img4 = rv.from_numpy(np.random.rand(50, 10, 10))
img5 = rv.from_numpy(np.random.rand(10, 50, 10))
img6 = rv.from_numpy(np.random.rand(10, 10, 50))
rs1 = rv.RadSlicer(img1, title="Image 1")
rs2 = rv.RadSlicer(img2, title="Image 2")
rs3 = rv.RadSlicer(img3, title="Image 3")
rs4 = rv.RadSlicer(img4, title="Image 4")
rs5 = rv.RadSlicer(img5, title="Image 5")
rs6 = rv.RadSlicer(img6, title="Image 6")
rsg = rv.RadSlicerGroup([rs1, rs2, rs3, rs4, rs5, rs6], rows=2, cols=3)
rsg.update_slider_heights(0.05)
rsg.display()The processing module of RadVis offers a set of functions to perform preprocessing tasks
The percentile_clipping function clips pixel intensities above and below percentile ranges
The noise_reduction function reduces the amount of noise in the image
The normalization function normalizes the pixel intensities of the image to a specified range.
The add_padding function adds padding evenly to match a target shape
Example usage of processing functions:
import radvis as rv
import numpy as np
# Loading image
image = rv.load_image(filepath='path/to/image.nii.gz')
# Applying processing functions
clipped_image = rv.processing.percentile_clipping(image, lower_percentile=0.25, upper_percentile=0.75)
filtered_image = rv.processing.noise_reduction(clipped_image, filter_size=1)
normalized_image = rv.processing.normalization(filtered_image, min_val=0, max_val=255)
padded_image = rv.processing.add_padding(normalized_image, target_shape=(128, 128, 128))
# Displaying processed image
slicer = rv.RadSlicer(padded_image, axis=0)
slicer.display()