-
Notifications
You must be signed in to change notification settings - Fork 6
Create Validate Visualize Deposit ISA to Metabolights Workflow
By Philippe Rocca-Serra and David Johnson (last updated: 2018-01-16)
Archiving raw acquisition data in a repository without ancillary description diminished the value of dataset. It makes it harder for people to understand and possibly reuse the said dataset. In this tutorial, we highlight how to make the moment of the experimental plan to generate experimental metadata capable of supporting long term deposition and unambiguous description of a Metabolomics Study. To do so, we will rely on a Galaxy tool powered by a new functionality of the ISA-API to generate ISA standard compliant tabular or json documents.
The following publications provide more information about ISA format and its applications:
- Sansone A-S; ...; (2012). isa tools Nat Genet. 2012 January 27; 44(2): 121–126.
The purpose of this tutorial is to learn how to take advantage of your study design information to quickly generate ISA compliant documents, validate them and deposit them to EMBL-EBI Metabolights.
The workflow is available at our PhenoMeNal public instance and on deployed PhenoMeNal Cloud Research Environments (CRE).
The tutorial (which you can follow either with the videos or with the accompanying text) shows you how to:
- use the Create ISA study Galaxy Tool.
- validate the resulting metadata document using ISA-Tab validator function,
- preregister or deposit the ISA formatted study to EMBL-Metabolights.
Note: if you are new to Galaxy we strongly advise you to first read the "Galaxy 101 - What is Galaxy?" tutorial which will help you get a better understanding about the basic concepts when running workflows in Galaxy.
On the welcome page of Galaxy you will find a menu item "Login or register". Please make sure you are logged in before proceeding to step two. If you don't have an account yet, you can use the registration form to request access. To get an account on the PhenoMeNal public instance please check this tutorial. If you proceed without an account you will not be able to import the workflow and run it.
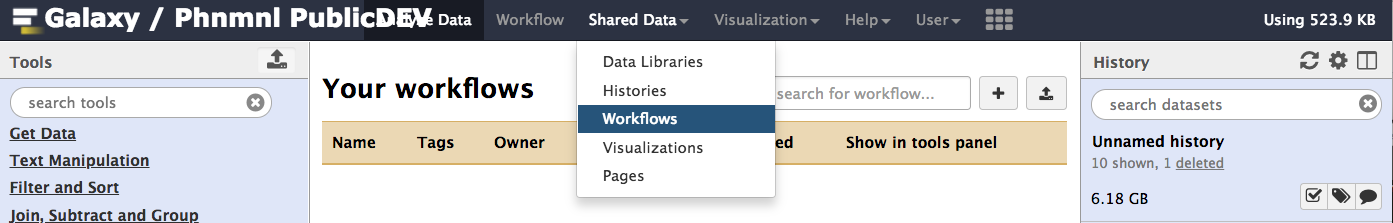
Once logged in, go the 'Shared Data' Menu in the toolbar and then go to the 'Workflow' menu item.
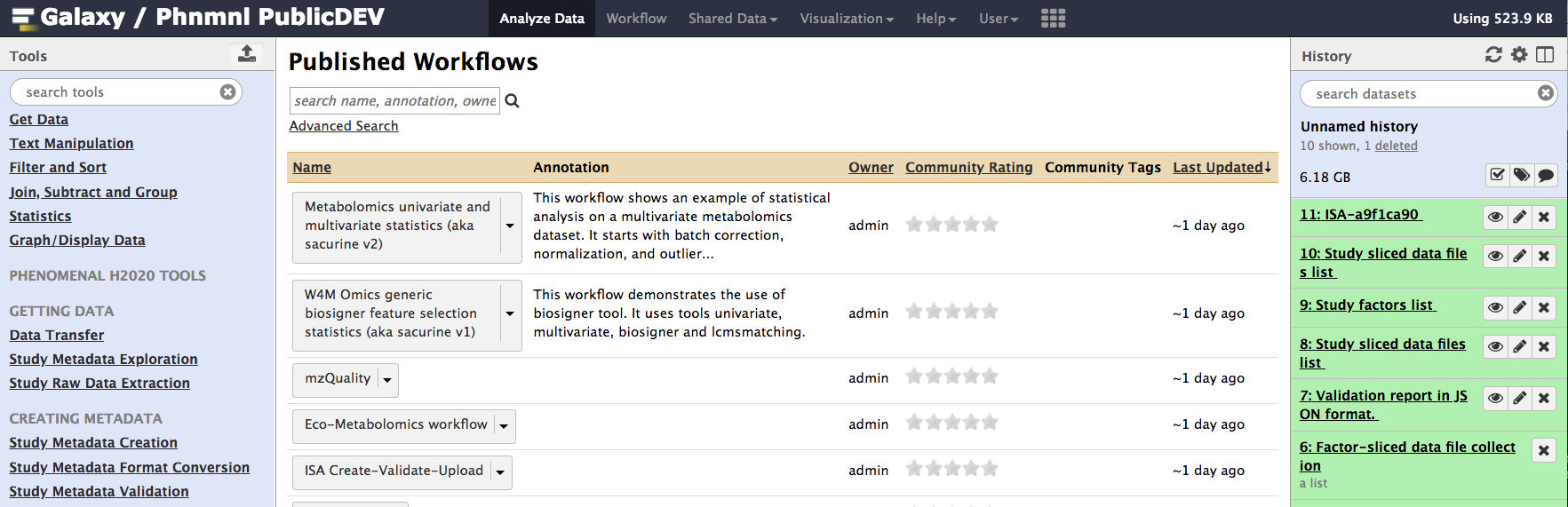
From the list of published workflows, select the 'ISA-Create-Validate-Upload' worflow and select 'Import' from the dropdown list of possible actions.
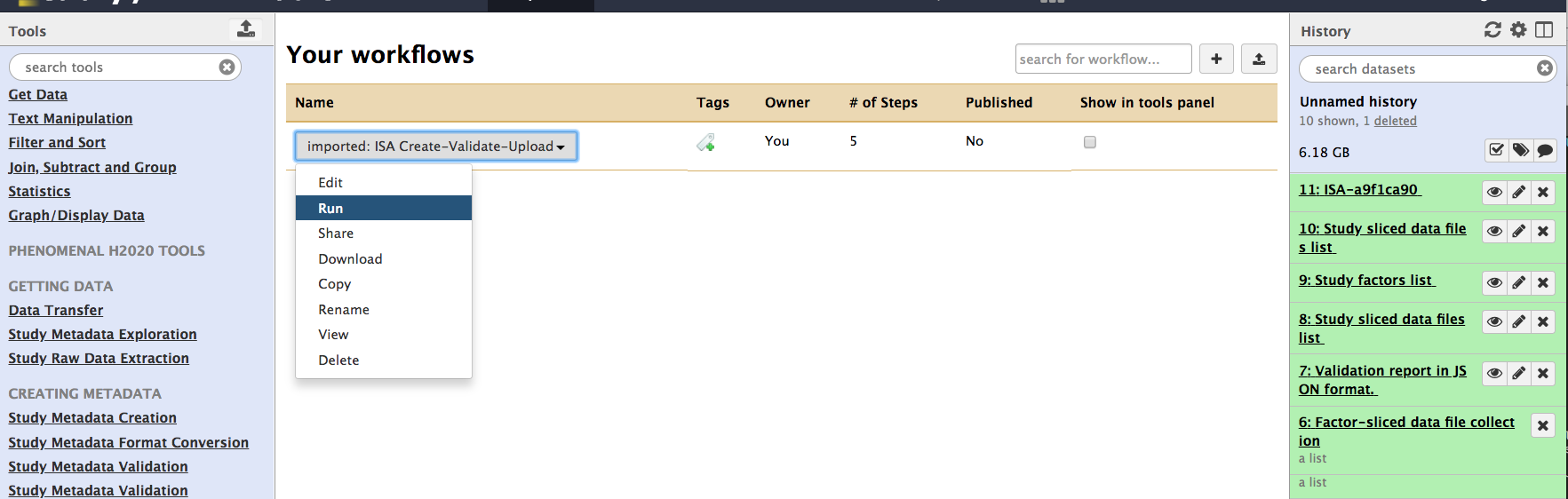
If all goes well, users are prompted with success message and they can select 'start using this workflow' to begin a full exploration of the component.
To do so, from this new page, navigate the ISA workflow and select 'Run'
If you are completely sure the default settings saved with this workflow are right for you, you can execute the workflow straight away.
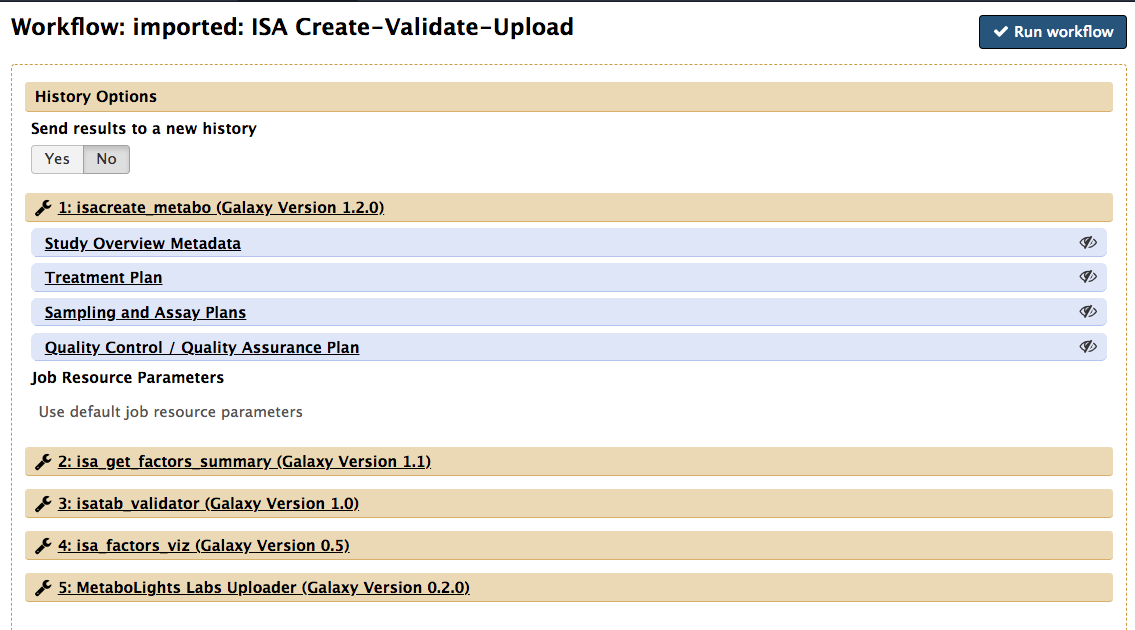
However, it is likely you'll want to use the saved workflow as a template. Therefore, you can take advantage of the edit function to alter the input parameters to match your needs.
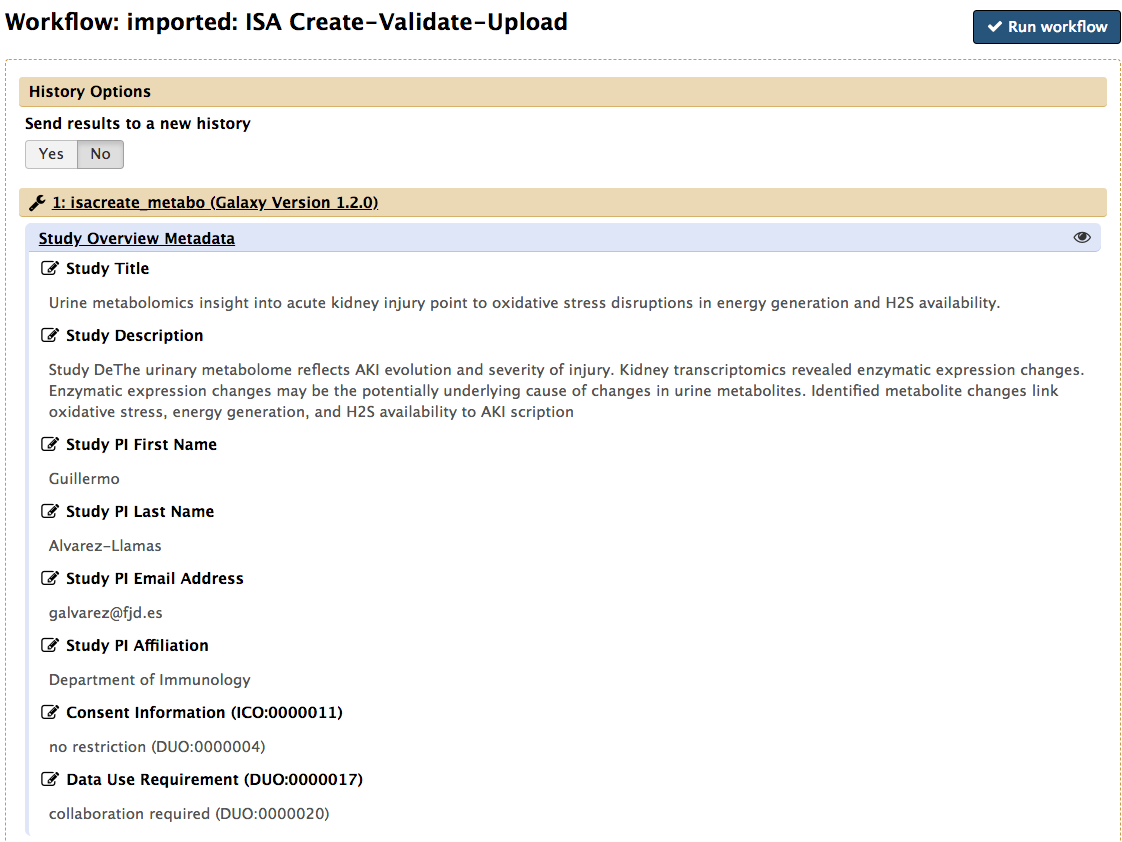
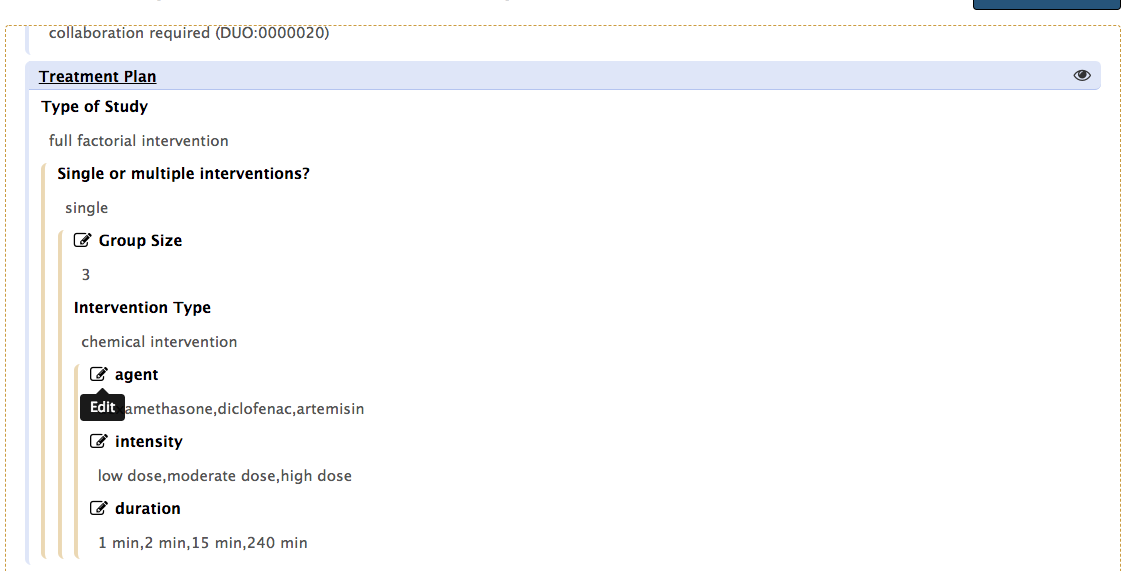
To do so, simply click the pencil icon to activate the edit functionality of any of the given field.
In order to upload an ISA archive to Metabolights, you'll need to create an account at the European database and generate an API key from your account profile. Once this done, simply copy the API key to the 'Upload' section of the workflow as shown below:
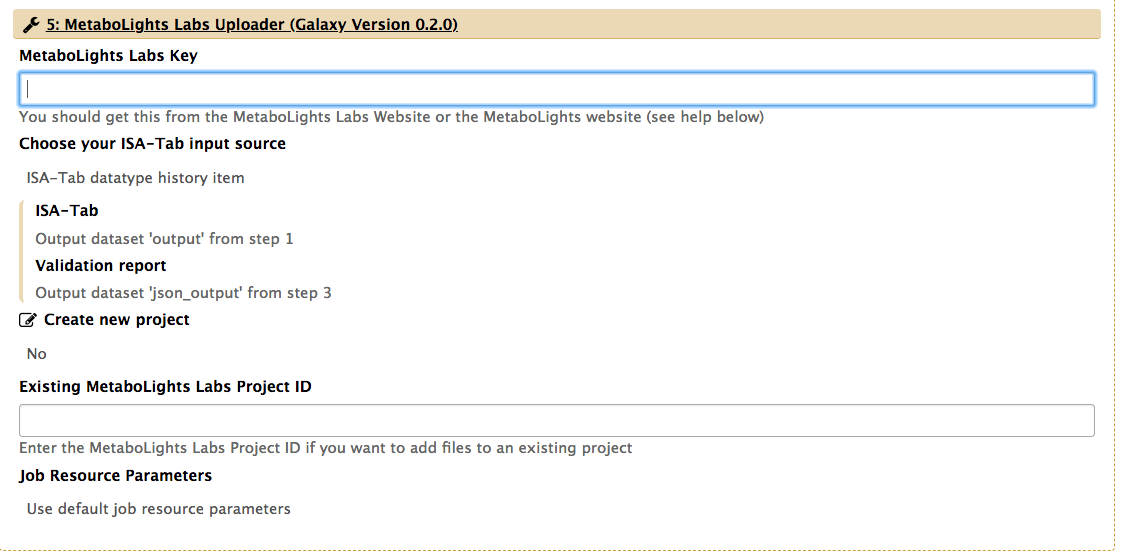
You are now all set to run a workflow that will create the study metadata descriptors based on study design information, execute the ISA syntactic validator and if the process is succesful, initiate the upload process to EBML-EBI Metabolights servers. Galaxy will indicate the job has been submitted
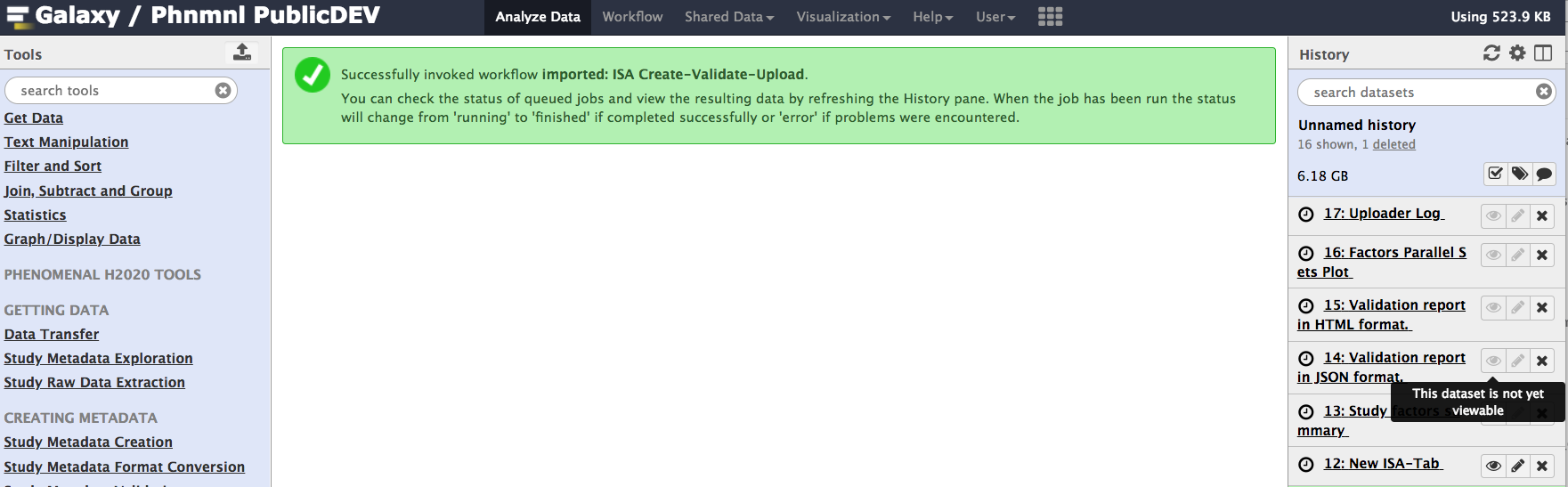
Assuming everything went well, let's check the output of this workflow. For optimal use of Galaxy visualization component, as a logged-in user, activate Galaxy Scratchpad. This will allow to spawn as many windows as you need and tile those side by side, thus providing different view points over the ISA formatted metadata.
This is can be to view the ISA metadata as html tables as in the top window, or as parallel coordinates showing the various study groups and the corresponding factor value combinations.
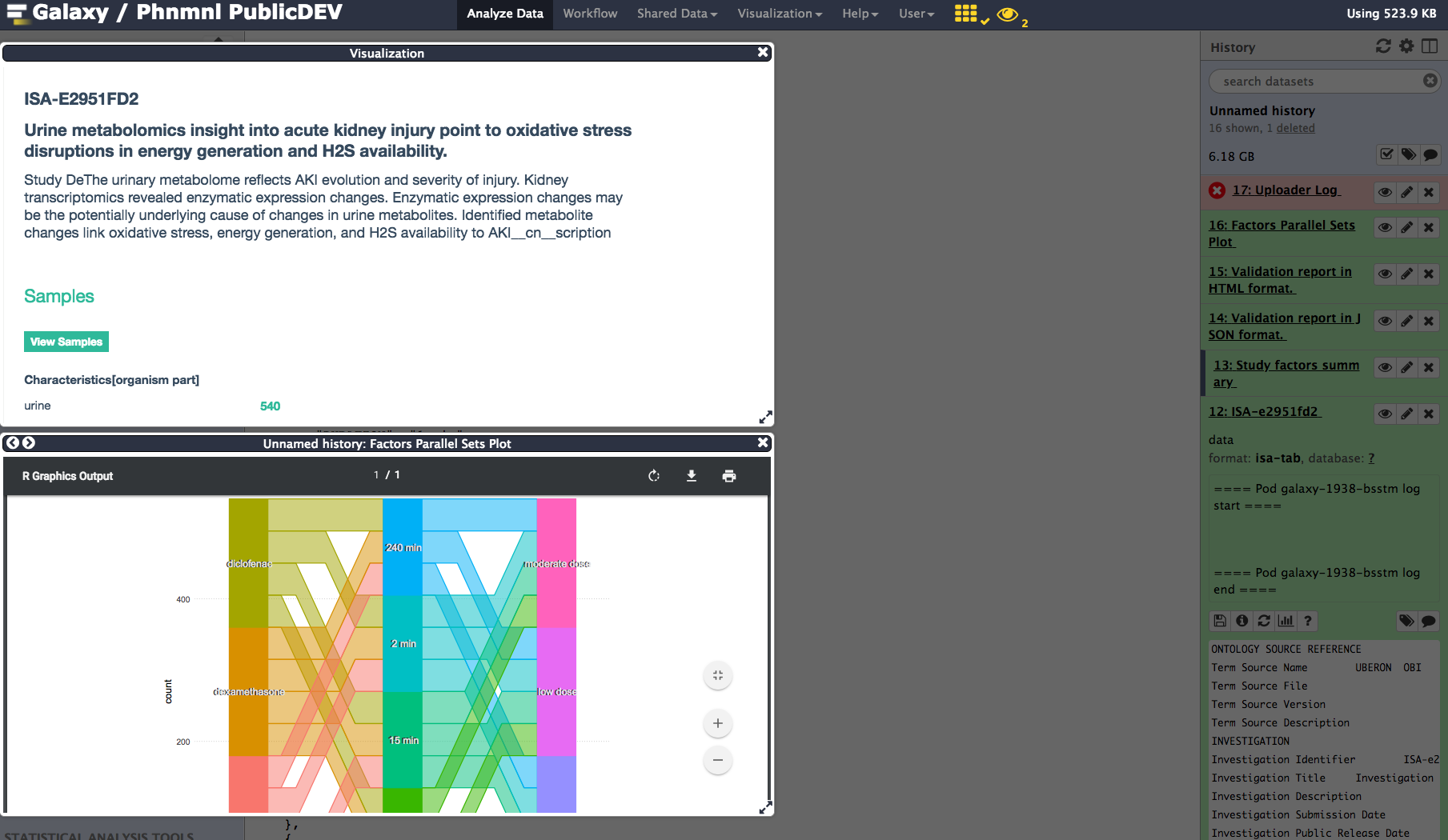
Another tile may be added to check the upload status to Metabolights. In this example, we show a report indicated validation success and upload being initiated.
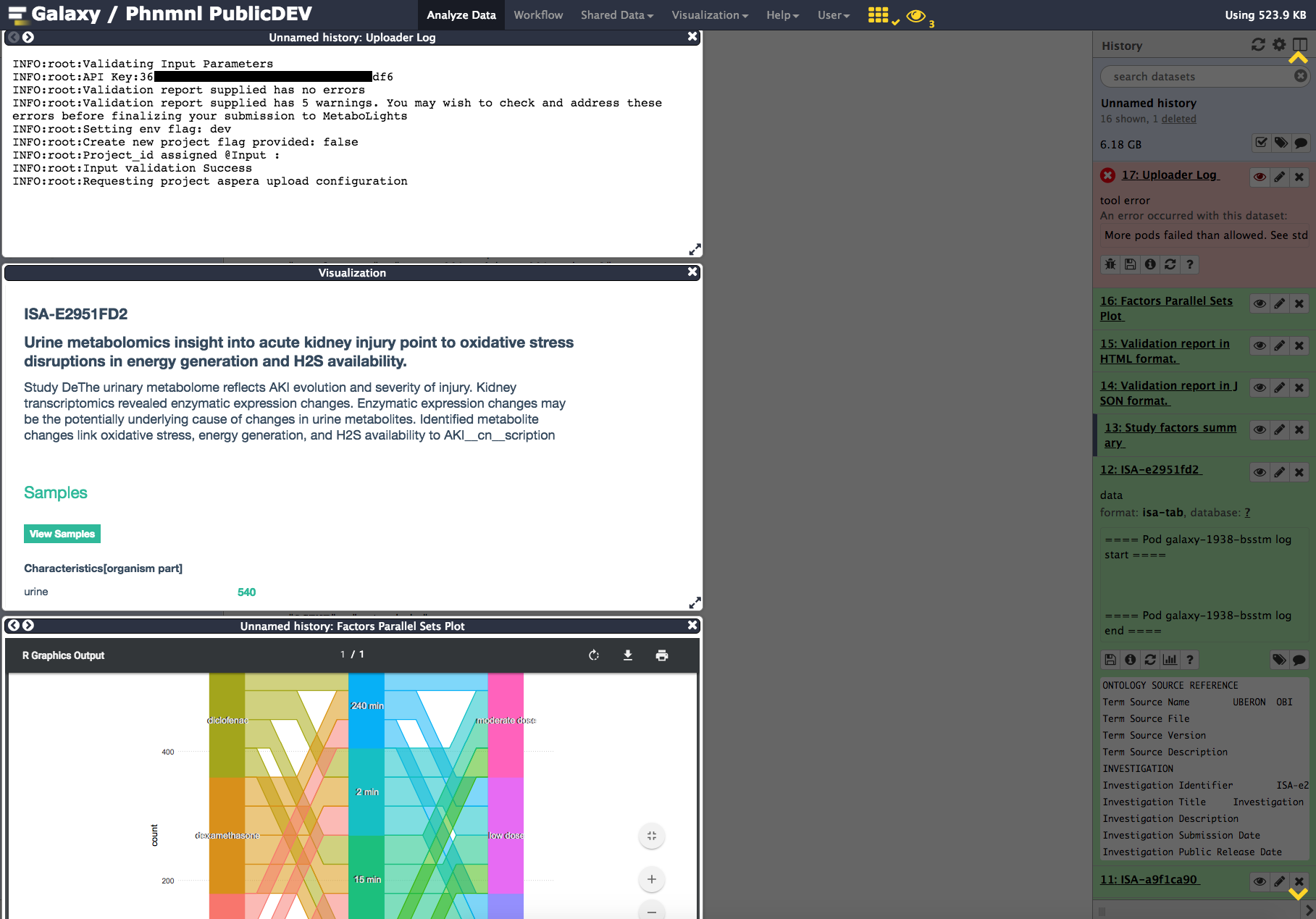
Running this ISA Metadata workflow is a very effective way to get acquainted with the various components supporting the ISA format and now available from Galaxy thanks to the PhenoMenAl project.
Once familiar with the tools and setup, you will be able to create,register,deposit and publish FAIR metabolomics datasets in no time.
Feel free to report any feedback to us by email or via the Online Feedback Form.
Other tutorials are available on our website (see the PhenoMeNal wiki).
All tools have been developed by the group of Susanna Sansone, from the University of Oxford.
All of these tools have been containerized by David Johnson and Philippe Rocca-Serra.
 |
Funded by the EC Horizon 2020 programme, grant agreement number 654241 |  |
|---|
